You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
134 Hilpert et al.<br />
wt A C D E F … … W Y<br />
MIDI AIDI CIDI<br />
DIDI<br />
EIDI<br />
FIDI<br />
WIDI<br />
YIDI<br />
MIDI MADI MCDI<br />
MDDI<br />
MEDI<br />
MFDI<br />
MWDI<br />
MYDI<br />
MIDI MIAI MICI<br />
MIDI MIEI MIFI<br />
MIWI<br />
MIYI<br />
MIDI MIDA MIDC MIDD MIDE MIDF MIDW MIDY<br />
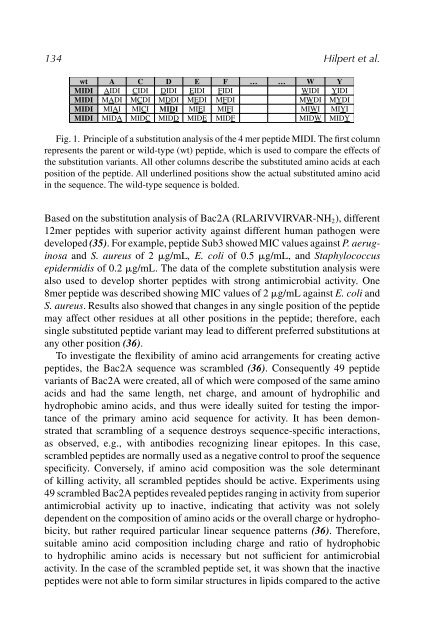
Fig. 1. Principle of a substitution analysis of the 4 mer peptide MIDI. The first column<br />
represents the parent or wild-type (wt) peptide, which is used to compare the effects of<br />
the substitution variants. All other columns describe the substituted amino acids at each<br />
position of the peptide. All underlined positions show the actual substituted amino acid<br />
in the sequence. The wild-type sequence is bolded.<br />
<strong>Based</strong> on the substitution analysis of Bac2A (RLARIVVIRVAR-NH2), different<br />
12mer peptides with superior activity against different human pathogen were<br />
developed (35). For example, peptide Sub3 showed MIC values against P. aeruginosa<br />
and S. aureus of 2 �g/mL, E.coliof 0.5 �g/mL, and Staphylococcus<br />
epidermidis of 0.2 �g/mL. The data of the complete substitution analysis were<br />
also used to develop shorter peptides with strong antimicrobial activity. One<br />
8mer peptide was described showing MIC values of 2 �g/mL against E. coli and<br />
S. aureus. Results also showed that changes in any single position of the peptide<br />
may affect other residues at all other positions in the peptide; therefore, each<br />
single substituted peptide variant may lead to different preferred substitutions at<br />
any other position (36).<br />
To investigate the flexibility of amino acid arrangements for creating active<br />
peptides, the Bac2A sequence was scrambled (36). Consequently 49 peptide<br />
variants of Bac2A were created, all of which were composed of the same amino<br />
acids and had the same length, net charge, and amount of hydrophilic and<br />
hydrophobic amino acids, and thus were ideally suited for testing the importance<br />
of the primary amino acid sequence for activity. It has been demonstrated<br />
that scrambling of a sequence destroys sequence-specific interactions,<br />
as observed, e.g., with antibodies recognizing linear epitopes. In this case,<br />
scrambled peptides are normally used as a negative control to proof the sequence<br />
specificity. Conversely, if amino acid composition was the sole determinant<br />
of killing activity, all scrambled peptides should be active. Experiments using<br />
49 scrambled Bac2A peptides revealed peptides ranging in activity from superior<br />
antimicrobial activity up to inactive, indicating that activity was not solely<br />
dependent on the composition of amino acids or the overall charge or hydrophobicity,<br />
but rather required particular linear sequence patterns (36). Therefore,<br />
suitable amino acid composition including charge and ratio of hydrophobic<br />
to hydrophilic amino acids is necessary but not sufficient for antimicrobial<br />
activity. In the case of the scrambled peptide set, it was shown that the inactive<br />
peptides were not able to form similar structures in lipids compared to the active