Tour-de-Force
Tour-de-Force
Tour-de-Force
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
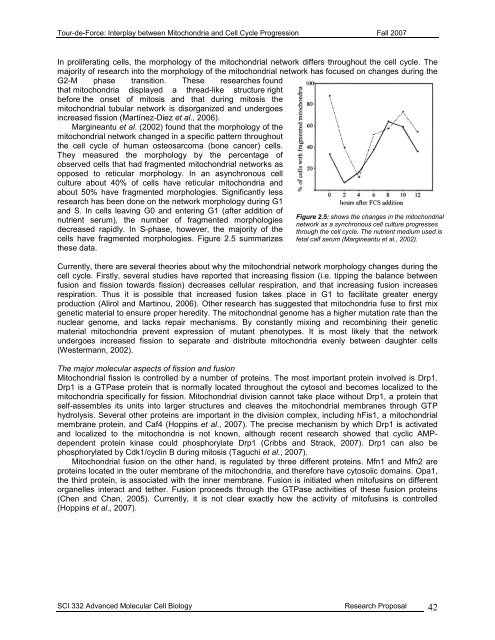
<strong>Tour</strong>-<strong>de</strong>-<strong>Force</strong>: Interplay between Mitochondria and Cell Cycle Progression Fall 2007In proliferating cells, the morphology of the mitochondrial network differs throughout the cell cycle. Themajority of research into the morphology of the mitochondrial network has focused on changes during theG2-M phase transition. These researches foundthat mitochondria displayed a thread-like structure rightbefore the onset of mitosis and that during mitosis themitochondrial tubular network is disorganized and un<strong>de</strong>rgoesincreased fission (Martínez-Diez et al., 2006).Margineantu et al. (2002) found that the morphology of themitochondrial network changed in a specific pattern throughoutthe cell cycle of human osteosarcoma (bone cancer) cells.They measured the morphology by the percentage ofobserved cells that had fragmented mitochondrial networks asopposed to reticular morphology. In an asynchronous cellculture about 40% of cells have reticular mitochondria andabout 50% have fragmented morphologies. Significantly lessresearch has been done on the network morphology during G1and S. In cells leaving G0 and entering G1 (after addition ofnutrient serum), the number of fragmented morphologies<strong>de</strong>creased rapidly. In S-phase, however, the majority of thecells have fragmented morphologies. Figure 2.5 summarizesthese data.Figure 2.5: shows the changes in the mitochondrialnetwork as a synchronous cell culture progressesthrough the cell cycle. The nutrient medium used isfetal calf serum (Margineantu et al., 2002).Currently, there are several theories about why the mitochondrial network morphology changes during thecell cycle. Firstly, several studies have reported that increasing fission (i.e. tipping the balance betweenfusion and fission towards fission) <strong>de</strong>creases cellular respiration, and that increasing fusion increasesrespiration. Thus it is possible that increased fusion takes place in G1 to facilitate greater energyproduction (Alirol and Martinou, 2006). Other research has suggested that mitochondria fuse to first mixgenetic material to ensure proper heredity. The mitochondrial genome has a higher mutation rate than thenuclear genome, and lacks repair mechanisms. By constantly mixing and recombining their geneticmaterial mitochondria prevent expression of mutant phenotypes. It is most likely that the networkun<strong>de</strong>rgoes increased fission to separate and distribute mitochondria evenly between daughter cells(Westermann, 2002).The major molecular aspects of fission and fusionMitochondrial fission is controlled by a number of proteins. The most important protein involved is Drp1.Drp1 is a GTPase protein that is normally located throughout the cytosol and becomes localized to themitochondria specifically for fission. Mitochondrial division cannot take place without Drp1, a protein thatself-assembles its units into larger structures and cleaves the mitochondrial membranes through GTPhydrolysis. Several other proteins are important in the division complex, including hFis1, a mitochondrialmembrane protein, and Caf4 (Hoppins et al., 2007). The precise mechanism by which Drp1 is activatedand localized to the mitochondria is not known, although recent research showed that cyclic AMP<strong>de</strong>pen<strong>de</strong>ntprotein kinase could phosphorylate Drp1 (Cribbs and Strack, 2007). Drp1 can also bephosphorylated by Cdk1/cyclin B during mitosis (Taguchi et al., 2007).Mitochondrial fusion on the other hand, is regulated by three different proteins. Mfn1 and Mfn2 areproteins located in the outer membrane of the mitochondria, and therefore have cytosolic domains. Opa1,the third protein, is associated with the inner membrane. Fusion is initiated when mitofusins on differentorganelles interact and tether. Fusion proceeds through the GTPase activities of these fusion proteins(Chen and Chan, 2005). Currently, it is not clear exactly how the activity of mitofusins is controlled(Hoppins et al., 2007).SCI 332 Advanced Molecular Cell Biology Research Proposal 42