Screening for cancer: are biomarkers of value?
Screening for cancer: are biomarkers of value?
Screening for cancer: are biomarkers of value?
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
– February/March 2011 28 Molecular diagnostics<br />
germline sequencing and insertion/deletion<br />
analysis <strong>of</strong> the MMR genes. If the<br />
tumour has been previously analysed by<br />
immunohistochemistry, targeted germline<br />
sequencing and insertion/deletion<br />
analysis <strong>of</strong> the gene corresponding to the<br />
missing MMR protein can be per<strong>for</strong>med.<br />
Pre-analytic and analytic<br />
considerations <strong>of</strong> molecular<br />
diagnostic methods<br />
Genetic testing <strong>of</strong> solid tissue specimens<br />
presents a unique challenge <strong>for</strong> the clinical<br />
laboratory. Samples vary widely in tissue<br />
quantity (e.g. fine needle biopsy vs. resection<br />
specimen), quality (e.g. fresh frozen<br />
vs. <strong>for</strong>malin-fixed <strong>for</strong> 72 hours), and heterogeneity<br />
<strong>of</strong> tumour and non-tumour<br />
tissue. Due to these factors, highly robust<br />
and sensitive methods that can detect a<br />
clinically significant variant present in a<br />
low proportion <strong>of</strong> the sample <strong>are</strong> needed.<br />
Traditional Sanger sequencing has a sensitivity<br />
<strong>of</strong> only ~15-20% to detect minor<br />
allele populations within a mixture,<br />
which can result in false negative results<br />
<strong>for</strong> KRAS testing [29]. Pyrosequencing<br />
and melting curve analysis generally have<br />
better sensitivity, at ~1-10% [29]. Newer<br />
methods that increase sensitivity further<br />
to ~0.1-1% include co-amplification at<br />
lower denaturation temperature (COLD)-<br />
PCR, and peptide nucleic acid (PNA)<br />
clamping [30, 31]. Such sensitive methods<br />
have been described in the literature <strong>for</strong><br />
KRAS [30-32] and BRAF [32] mutation<br />
detection in CRC, and have been shown<br />
to improve diagnostic accuracy. Additional<br />
techniques which could allow ultrasensitive<br />
detection <strong>of</strong> somatic mutations<br />
include next-generation sequencing [33]<br />
and limiting-dilution-PCR [34]. Importantly,<br />
with the increased ability to detect<br />
minor mutant populations within the<br />
sample, it will be necessary to determine<br />
the clinically relevant threshold in addition<br />
to the absolute analytical threshold.<br />
Conclusions and<br />
recommendations<br />
Marked improvements in our understanding<br />
<strong>of</strong> CRC biology over the last few years<br />
have led to the discovery <strong>of</strong> several genetic<br />
<strong>biomarkers</strong> that guide treatment and<br />
establish prognosis <strong>of</strong> patients with CRC.<br />
Although many <strong>biomarkers</strong> have been<br />
studied, currently only KRAS, BRAF, and<br />
MSI <strong>are</strong> sufficiently validated to support<br />
routine clinical use. Cases in which MSI is<br />
suspected based on clinicopathologic data<br />
should be tested. If MSI is demonstrated,<br />
the distinction between Lynch syndrome<br />
and sporadic CRC is fundamental to determine<br />
the necessity <strong>of</strong> genetic counselling<br />
and close oncologic surveillance. Sporadic<br />
CRC cases should be evaluated <strong>for</strong> KRAS<br />
and possibly BRAF if anti-EGFR therapy is<br />
an option. In the future, testing the PI3K<br />
pathway genes PIK3CA and PTEN may<br />
help define the population which is most<br />
likely to benefit from the expensive anti-<br />
EGFR treatment. The per<strong>for</strong>mance <strong>of</strong> the<br />
molecular methods used <strong>for</strong> the above<br />
determinations needs to be considered<br />
and clearly communicated so that patients<br />
receive the most accurate and meaningful<br />
in<strong>for</strong>mation, and do not over-interpret the<br />
laboratory data.<br />
As the era <strong>of</strong> molecular oncology and<br />
personalised medicine unfolds, many<br />
changes in the way <strong>cancer</strong> is currently<br />
diagnosed and treated <strong>are</strong> expected to<br />
occur. Laboratory directors and staff alike<br />
have the responsibility <strong>of</strong> keeping up with<br />
the pace <strong>of</strong> change to continue <strong>of</strong>fering<br />
the best diagnostic testing to clinicians<br />
and patients.<br />
References<br />
1. Karsa LV et al. Best Pract Res Clin Gastroenterol<br />
2010; 24(4): 381-396.<br />
2. Boland CR et al. Gastroenterology 2010;<br />
138(6):2073-2087.e2073.<br />
3. Vilar E et al. Nat Rev Clin Oncol 2010; 7(3):153-162.<br />
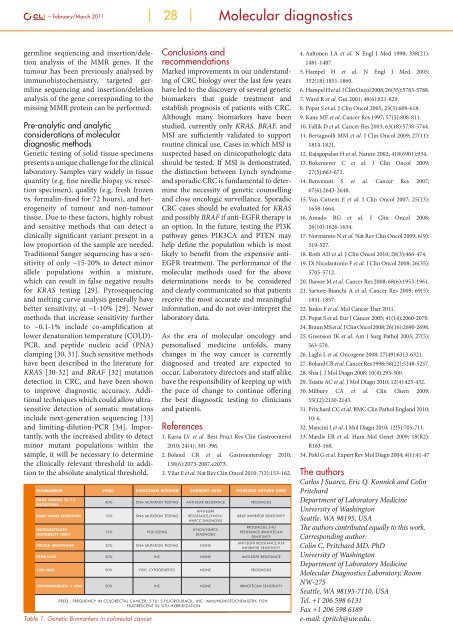
Biomarker Freq. Detection Method Current Uses Possible Future Uses<br />
KRAS codon 12/13<br />
mutations<br />
40% DNA mutation testing Anti-EGFR resistance Prognosis<br />
BRAF V600E mutation 10% DNA mutation testing<br />
Microsatellite<br />
instability (MSI)<br />
15% PCR/sizing<br />
Anti-EGFR<br />
resistance,Lynch/<br />
HNPCC diagnosis<br />
Lynch/HNPCC<br />
diagnosis<br />
PIK3CA mutations 20% DNA mutation testing None<br />
BRAF inhibitor sensitivity<br />
Prognosis,5-FU<br />
resistance,Irinotecan<br />
sensitivity<br />
Anti-EGFR resistance,PI3K<br />
inhibitor sensitivity<br />
PTEN loss 30% IHC None Anti-EGFR resistance<br />
18q loss 50% FISH, cytogenetics None Prognosis<br />
Topoisomerase 1 low 50% IHC None Irinotecan sensitivity<br />
Freq.: Frequency in colorectal <strong>cancer</strong>; 5-FU: 5-fluorouracil; IHC: Immunohistochemistry; FISH:<br />
Fluorescent in situ hybridization<br />
Table 1. Genetic Biomarkers in colorectal <strong>cancer</strong>.<br />
4. Aaltonen LA et al. N Engl J Med 1998; 338(21):<br />
1481-1487.<br />
5. Hampel H et al. N Engl J Med 2005;<br />
352(18):1851-1860.<br />
6. Hampel H et al. J Clin Oncol 2008; 26(35):5783-5788.<br />
7. Ward R et al. Gut 2001; 48(6):821-829.<br />
8. Popat S et al. J Clin Oncol 2005; 23(3):609-618.<br />
9. Kane MF et al. Cancer Res 1997; 57(5):808-811.<br />
10. Fallik D et al. Cancer Res 2003; 63(18):5738-5744.<br />
11. Bertagnolli MM et al. J Clin Oncol 2009; 27(11):<br />
1814-1821.<br />
12. Rajagopalan H et al. Nature 2002; 418(6901):934.<br />
13. Bokemeyer C et al. J Clin Oncol 2009;<br />
27(5):663-671.<br />
14. Benvenuti S et al. Cancer Res 2007;<br />
67(6):2643-2648.<br />
15. Van Cutsem E et al. J Clin Oncol 2007; 25(13):<br />
1658-1664.<br />
16. Amado RG et al. J Clin Oncol 2008;<br />
26(10):1626-1634.<br />
17. Normanno N et al. Nat Rev Clin Oncol 2009; 6(9):<br />
519-527.<br />
18. Roth AD et al. J Clin Oncol 2010; 28(3):466-474.<br />
19. Di Nicolantonio F et al. J Clin Oncol 2008; 26(35):<br />
5705-5712.<br />
20. Jhawer M et al. Cancer Res 2008; 68(6):1953-1961.<br />
21. Sartore-Bianchi A et al. Cancer Res 2009; 69(5):<br />
1851-1857.<br />
22. Janku F et al. Mol Cancer Ther 2011.<br />
23. Popat S et al. Eur J Cancer 2005; 41(14):2060-2070.<br />
24. Braun MS et al. J Clin Oncol 2008; 26(16):2690-2698.<br />
25. Greenson JK et al. Am J Surg Pathol 2003; 27(5):<br />
563-570.<br />
26. Laghi L et al. Oncogene 2008; 27(49):6313-6321.<br />
27. Boland CR et al. Cancer Res 1998; 58(22):5248-5257.<br />
28. Shia J. J Mol Diagn 2008; 10(4):293-300.<br />
29. Tsiatis AC et al. J Mol Diagn 2010; 12(4):425-432.<br />
30. Milbury CA et al. Clin Chem 2009;<br />
55(12):2130-2143.<br />
31. Pritchard CC et al. BMC Clin Pathol England 2010;<br />
10: 6.<br />
32. Mancini I et al. J Mol Diagn 2010; 12(5):705-711.<br />
33. Mardis ER et al. Hum Mol Genet 2009; 18(R2):<br />
R163-168.<br />
34. Pohl G et al. Expert Rev Mol Diagn 2004; 4(1):41-47<br />
The authors<br />
Carlos J Su<strong>are</strong>z, Eric Q. Konnick and Colin<br />
Pritchard<br />
Department <strong>of</strong> Laboratory Medicine<br />
University <strong>of</strong> Washington<br />
Seattle, WA 98195, USA<br />
The authors contributed equally to this work.<br />
Corresponding author:<br />
Colin C. Pritchard MD, PhD<br />
University <strong>of</strong> Washington<br />
Department <strong>of</strong> Laboratory Medicine<br />
Molecular Diagnostics Laboratory, Room<br />
NW-275<br />
Seattle, WA 98195-7110, USA<br />
Tel. +1 206 598 6131<br />
Fax +1 206 598 6189<br />
e-mail: cpritch@uw.edu.