Real time PCR - European Pharmaceutical Review
Real time PCR - European Pharmaceutical Review
Real time PCR - European Pharmaceutical Review
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
SUBSCRIBE<br />
ISSUE<br />
2009 q<strong>PCR</strong><br />
assays. Tests were developed to<br />
detect mycotoxins, T-toxin,<br />
phytoestrogens such as Coumestrol<br />
and several other contaminants,<br />
particularly in food.<br />
Quantification of RNA<br />
This obviously is the area that has<br />
most immediately beneficiated from<br />
real <strong>time</strong> <strong>PCR</strong>. It is well accepted that<br />
measures of gene expression is now<br />
quantitative rather than semiquantitative.<br />
Two methods exist, one<br />
absolute comparing gene expression<br />
to known standards and a second,<br />
relative to an endogenous reference<br />
(usually a house keeping gene).<br />
Outside its plain use for gene<br />
expression studies (in cells in<br />
response to stimuli for example), real<br />
<strong>time</strong> <strong>PCR</strong> has been instrumental<br />
notably in transcriptome studies for<br />
confirming micro-array work, when<br />
limited amount of material is available<br />
(small tissue biopsy studies), or for<br />
large studies necessitation high<br />
thorough-put and automatism with<br />
<strong>PCR</strong> robots. This has notably<br />
warranted the development and<br />
success of the micro-fluidic card<br />
systems. It was recently argued 7 that<br />
advances in the tumour biology have<br />
now provided molecular targets for<br />
diagnosing and treating cancer.<br />
Clustering algorithms have revealed a<br />
molecular diversity that forms a new<br />
taxonomy with diagnostic, prognostic<br />
and possibly therapeutic significance.<br />
The challenge in pathology today is<br />
to develop and implement such<br />
molecular classifications in routine<br />
clinical practice. This requires<br />
objective, robust, and cost-effective<br />
techniques and real-<strong>time</strong> <strong>PCR</strong> offers<br />
such attractive features.<br />
Discrimination assay<br />
Genotyping using end point assays or<br />
melting curves analysis assays<br />
provides rapid results (raw biological<br />
samples to SNP genotyping results in<br />
less than one hour), highly accurate in<br />
virtually any sample (low amount of<br />
material or poor quality samples) and<br />
an increasing number of publications<br />
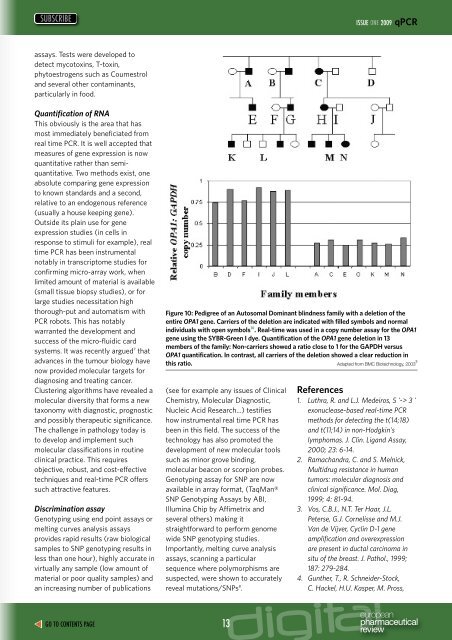
Figure 10: Pedigree of an Autosomal Dominant blindness family with a deletion of the<br />
entire OPA1 gene. Carriers of the deletion are indicated with filled symbols and normal<br />
individuals with open symbols 10 . <strong>Real</strong>-<strong>time</strong> was used in a copy number assay for the OPA1<br />
gene using the SYBR-Green I dye. Quantification of the OPA1 gene deletion in 13<br />
members of the family: Non-carriers showed a ratio close to 1 for the GAPDH versus<br />
OPA1 quantification. In contrast, all carriers of the deletion showed a clear reduction in<br />
this ratio. Adapted from BMC Biotechnology, 2003 5<br />
(see for example any issues of Clinical<br />
Chemistry, Molecular Diagnostic,<br />
Nucleic Acid Research…) testifies<br />
how instrumental real <strong>time</strong> <strong>PCR</strong> has<br />
been in this field. The success of the<br />
technology has also promoted the<br />
development of new molecular tools<br />
such as minor grove binding,<br />
molecular beacon or scorpion probes.<br />
Genotyping assay for SNP are now<br />
available in array format, (TaqMan®<br />
SNP Genotyping Assays by ABI,<br />
Illumina Chip by Affimetrix and<br />
several others) making it<br />
straightforward to perform genome<br />
wide SNP genotyping studies.<br />
Importantly, melting curve analysis<br />
assays, scanning a particular<br />
sequence where polymorphisms are<br />
suspected, were shown to accurately<br />
reveal mutations/SNPs 8 .<br />
References<br />
1. Luthra, R. and L.J. Medeiros, 5 '-> 3 '<br />
exonuclease-based real-<strong>time</strong> <strong>PCR</strong><br />
methods for detecting the t(14;18)<br />
and t(11;14) in non-Hodgkin's<br />
lymphomas. J. Clin. Ligand Assay,<br />
2000; 23: 6-14.<br />
2. Ramachandra, C. and S. Melnick,<br />
Multidrug resistance in human<br />
tumors: molecular diagnosis and<br />
clinical significance. Mol. Diag,<br />
1999; 4: 81-94.<br />
3. Vos, C.B.J., N.T. Ter Haar, J.L.<br />
Peterse, G.J. Cornelisse and M.J.<br />
Van de Vijver, Cyclin D-1 gene<br />
amplification and overexpression<br />
are present in ductal carcinoma in<br />
situ of the breast. J. Pathol., 1999;<br />
187: 279-284.<br />
4. Gunther, T., R. Schneider-Stock,<br />
C. Hackel, H.U. Kasper, M. Pross,<br />
GO TO CONTENTS PAGE 13