Real time PCR - European Pharmaceutical Review
Real time PCR - European Pharmaceutical Review
Real time PCR - European Pharmaceutical Review
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
q<strong>PCR</strong> ISSUE 2009<br />
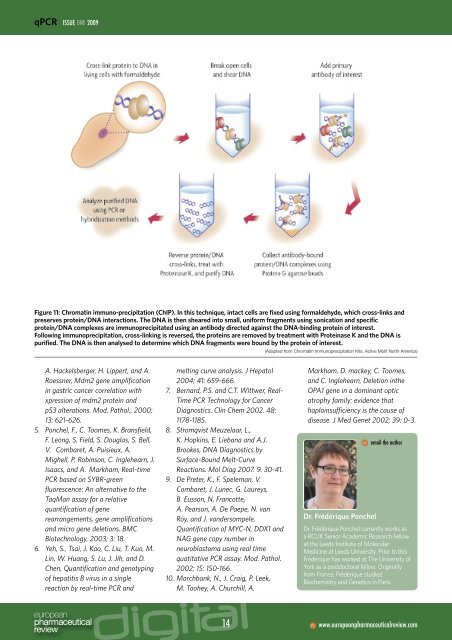
Figure 11: Chromatin immuno-precipitation (ChIP). In this technique, intact cells are fixed using formaldehyde, which cross-links and<br />
preserves protein/DNA interactions. The DNA is then sheared into small, uniform fragments using sonication and specific<br />
protein/DNA complexes are immunoprecipitated using an antibody directed against the DNA-binding protein of interest.<br />
Following immunoprecipitation, cross-linking is reversed, the proteins are removed by treatment with Proteinase K and the DNA is<br />
purified. The DNA is then analysed to determine which DNA fragments were bound by the protein of interest.<br />
(Adapted from Chromatin Immunoprecipitation Kits, Active Motif North America)<br />
A. Hackelsberger, H. Lippert, and A.<br />
Roessner, Mdm2 gene amplification<br />
in gastric cancer correlation with<br />
xpression of mdm2 protein and<br />
p53 alterations. Mod. Pathol., 2000;<br />
13: 621-626.<br />
5. Ponchel, F., C. Toomes, K. Bransfield,<br />
F. Leong, S. Field, S. Douglas, S. Bell,<br />
V. Combaret, A. Puisieux, A.<br />
Mighell, P. Robinson, C. Inglehearn, J.<br />
Isaacs, and A. Markham, <strong>Real</strong>-<strong>time</strong><br />
<strong>PCR</strong> based on SYBR-green<br />
fluorescence: An alternative to the<br />
TaqMan assay for a relative<br />
quantification of gene<br />
rearrangements, gene amplifications<br />
and micro gene deletions. BMC<br />
Biotechnology, 2003; 3: 18.<br />
6. Yeh, S., Tsai, J. Kao, C. Liu, T. Kuo, M.<br />
Lin, W. Huang, S. Lu, J. Jih, and D.<br />
Chen, Quantification and genotyping<br />
of hepatitis B virus in a single<br />
reaction by real-<strong>time</strong> <strong>PCR</strong> and<br />
melting curve analysis. J Hepatol<br />
2004; 41: 659-666.<br />
7. Bernard, P.S. and C.T. Wittwer, <strong>Real</strong>-<br />
Time <strong>PCR</strong> Technology for Cancer<br />
Diagnostics. Clin Chem 2002. 48:<br />
1178-1185.<br />
8. Stromqvist Meuzelaar, L.,<br />
K. Hopkins, E. Liebana and A.J.<br />
Brookes, DNA Diagnostics by<br />
Surface-Bound Melt-Curve<br />
Reactions. Mol Diag 2007. 9. 30-41.<br />
9. De Preter, K., F. Speleman, V.<br />
Combaret, J. Lunec, G. Laureys,<br />
B. Eusson, N. Francotte,<br />
A. Pearson, A. De Paepe, N. van<br />
Roy, and J. vandersompele,<br />
Quantification of MYC-N, DDX1 and<br />
NAG gene copy number in<br />
neuroblastoma using real <strong>time</strong><br />
quatitative <strong>PCR</strong> assay. Mod. Pathol.<br />
2002; 15: 150-166.<br />
10. Marchbank, N., J. Craig, P. Leek,<br />
M. Toohey, A. Churchill, A.<br />
Markham, D. mackey, C. Toomes,<br />
and C. Inglehearn, Deletion inthe<br />
OPA1 gene in a dominant optic<br />
atrophy family: evidence that<br />
haploinsufficiency is the cause of<br />
disease. J Med Genet 2002; 39: 0-3.<br />
Dr. Frédérique Ponchel<br />
Dr. Frédérique Ponchel currently works as<br />
a RCUK Senior Academic Research Fellow<br />
at the Leeds Institute of Molecular<br />
Medicine at Leeds University. Prior to this<br />
Frédérique has worked at The University of<br />
York as a postdoctoral fellow. Originally<br />
from France, Frédérique studied<br />
Biochemistry and Genetics in Paris.<br />
@<br />
email the author<br />
14<br />
www.europeanpharmaceuticalreview.com