Real time PCR - European Pharmaceutical Review
Real time PCR - European Pharmaceutical Review
Real time PCR - European Pharmaceutical Review
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
q<strong>PCR</strong> ISSUE 2009<br />
<strong>Real</strong> <strong>time</strong> <strong>PCR</strong><br />
Dr. Frédérique Ponchel, Senior Research Fellow, Leeds Institute of Molecular Medicine, University of Leeds<br />
Polymerase Chain Reaction (<strong>PCR</strong>) has revolutionised molecular biology, and is now a wellstandardised<br />
procedure and its applications are wide. <strong>Real</strong> <strong>time</strong> <strong>PCR</strong> is a more recent development<br />
of the initial <strong>PCR</strong> reaction, which has provided a solution to the limitation of standard <strong>PCR</strong> with<br />
regards to quantification. By using fluorescence, which allows the quantification of <strong>PCR</strong> products at<br />
each cycle of the reaction, real <strong>time</strong> <strong>PCR</strong> avoids the “plateau” effect of standard <strong>PCR</strong>. Two main<br />
approaches have been developed, the TaqMAn assay and the SYBR-Green I dye. Alongside real<strong>time</strong><br />
<strong>PCR</strong> as such, an “end point assay” has also evolved, most often referred to as allelic<br />
discrimination, allowing the detection of a specific sequence (i.e. genetic polymorphism). The<br />
technology is now widely used for DNA and RNA quantification as well as genetic polymorphism<br />
detection. It can be applied to the detection and quantification of plain mRNA, for splice variant,<br />
alternative promoter usage, allele specific expression etc. For DNA, gene amplification, gene<br />
deletion or rearrangement, allelic copy number can be measured. It is also widely used for the<br />
detection of pathogens or contaminants in food or other types of products.<br />
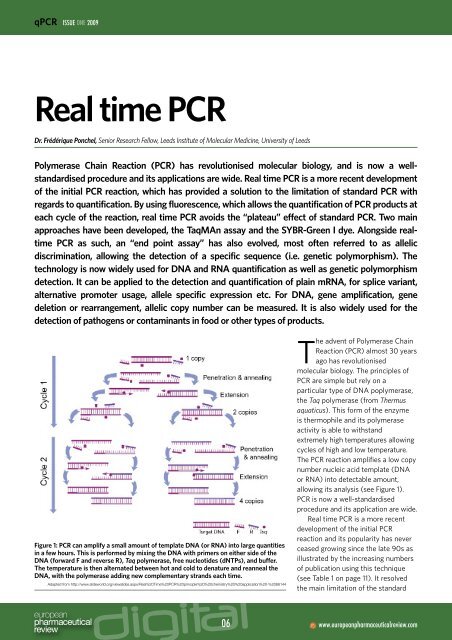
Figure 1: <strong>PCR</strong> can amplify a small amount of template DNA (or RNA) into large quantities<br />
in a few hours. This is performed by mixing the DNA with primers on either side of the<br />
DNA (forward F and reverse R), Taq polymerase, free nucleotides (dNTPs), and buffer.<br />
The temperature is then alternated between hot and cold to denature and reanneal the<br />
DNA, with the polymerase adding new complementary strands each <strong>time</strong>.<br />
Adapted from http://www.slideworld.org/viewslides.aspx/<strong>Real</strong>%20Time%20<strong>PCR</strong>%20principle%20%20chemistry%20%20application%20-%2066144<br />
The advent of Polymerase Chain<br />
Reaction (<strong>PCR</strong>) almost 30 years<br />
ago has revolutionised<br />
molecular biology. The principles of<br />
<strong>PCR</strong> are simple but rely on a<br />
particular type of DNA poplymerase,<br />
the Taq polymerase (from Thermus<br />
aquaticus). This form of the enzyme<br />
is thermophile and its polymerase<br />
activity is able to withstand<br />
extremely high temperatures allowing<br />
cycles of high and low temperature.<br />
The <strong>PCR</strong> reaction amplifies a low copy<br />
number nucleic acid template (DNA<br />
or RNA) into detectable amount,<br />
allowing its analysis (see Figure 1).<br />
<strong>PCR</strong> is now a well-standardised<br />
procedure and its application are wide.<br />
<strong>Real</strong> <strong>time</strong> <strong>PCR</strong> is a more recent<br />
development of the initial <strong>PCR</strong><br />
reaction and its popularity has never<br />
ceased growing since the late 90s as<br />
illustrated by the increasing numbers<br />
of publication using this technique<br />
(see Table 1 on page 11). It resolved<br />
the main limitation of the standard<br />
06<br />
www.europeanpharmaceuticalreview.com