Gene Variant Report issued
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
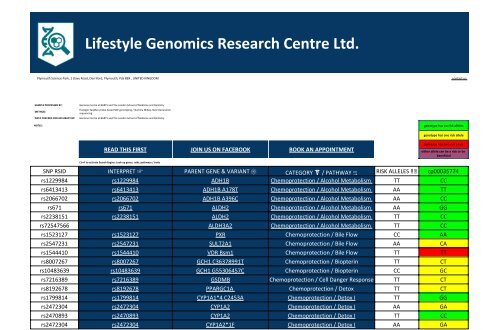
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1229984 rs1229984 ADH1B Chemoprotection / Alcohol Metabolism TT CC<br />
rs6413413 rs6413413 ADH1B A178T Chemoprotection / Alcohol Metabolism AA TT<br />
rs2066702 rs2066702 ADH1B A396C Chemoprotection / Alcohol Metabolism AA CC<br />
rs671 rs671 ALDH2 Chemoprotection / Alcohol Metabolism AA GG<br />
rs2238151 rs2238151 ALDH2 Chemoprotection / Alcohol Metabolism TT CC<br />
rs72547566 ALDH3A2 Chemoprotection / Alcohol Metabolism TT CC<br />
rs1523127 rs1523127 PXR Chemoprotection / Bile Flow CC AA<br />
rs2547231 rs2547231 SULT2A1 Chemoprotection / Bile Flow AA CA<br />
rs1544410 rs1544410 VDR Bsm1 Chemoprotection / Bile Flow TT TT<br />
rs8007267 rs8007267 GCH1 C36378991T Chemoprotection / Biopterin TT CT<br />
rs10483639 rs10483639 GCH1 G55306457C Chemoprotection / Biopterin CC GC<br />
rs7216389 rs7216389 GSDMB Chemoprotection / Cell Danger Response TT CT<br />
rs8192678 rs8192678 PPARGC1A Chemoprotection / Detox TT CT<br />
rs1799814 rs1799814 CYP1A1*4 C2453A Chemoprotection / Detox I TT GG<br />
rs2472304 rs2472304 CYP1A2 Chemoprotection / Detox I AA GA<br />
rs2470893 rs2470893 CYP1A2 Chemoprotection / Detox I TT CC<br />
rs2472304 rs2472304 CYP1A2*1F Chemoprotection / Detox I AA GA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs762551 rs762551 CYP1A2*1F C164A Chemoprotection / Detox I CC CA<br />
rs1056836 Hydroxylation of Estrogens CYP1B1 L432V Chemoprotection / Detox I CC GG<br />
rs1800440<br />
Estrogen Metabolism. Converts<br />
estradiol to 4-OH-estradiol<br />
CYP1B1 N453S Chemoprotection / Detox I TT TC<br />
rs9282671 Hydroxylation of Estrogens CYP1B1 T241A Chemoprotection / Detox I AA AA<br />
rs1801272<br />
Xenobiotics, nicotine, drugs<br />
metabolism. Works with CYP2B6<br />
CYP2A6*2 A1799T Chemoprotection / Detox I TT AA<br />
rs6583954 rs6583954 CYP2C19 Chemoprotection / Detox I TT CC<br />
rs4986894 rs4986894 CYP2C19 T98C Chemoprotection / Detox I TT TT<br />
rs41291556 rs41291556 CYP2C19*8 T358C Chemoprotection / Detox I CC TT<br />
rs2185570 rs2185570 CYP2C9 Chemoprotection / Detox I CC TT<br />
rs28371685 rs28371685 CYP2C9 Chemoprotection / Detox I TT CC<br />
rs9332239 rs9332239 CYP2C9*12 1465C>T Chemoprotection / Detox I TT CC<br />
rs1799853 rs1799853 CYP2C9*2 Chemoprotection / Detox I CT CC<br />
rs1057910 rs1057910 CYP2C9*3 Chemoprotection / Detox I CC AC<br />
rs9332131 rs9332131 CYP2C9*6 818delA Chemoprotection / Detox I DD AA<br />
rs7900194 rs7900194 CYP2C9*8 Chemoprotection / Detox I AA GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs28371706 rs28371706 CYP2D6*17 T107I Chemoprotection / Detox I AA GG<br />
rs5030867 rs5030867 CYP2D6*7 Chemoprotection / Detox I GG TT<br />
rs2740574<br />
Androgens, Xenobiotics, drugs<br />
metabolism, testosterone oxidation.<br />
CYP3A4*1B A392G Chemoprotection / Detox I GG TT<br />
rs3741049 rs3741049 ACAT1 G22670A Chemoprotection / Detox II AA GG<br />
rs2273684 rs2273684 GSS Chemoprotection / Detox II TT GG<br />
rs6088659 rs6088659 GSS A5997G Chemoprotection / Detox II TT CC<br />
rs1138272 rs1138272 GSTP1 Chemoprotection / Detox II TT CC<br />
rs1695 rs1695 GSTP1 Chemoprotection / Detox II GG AA<br />
rs1799929 rs1799929 NAT2 Chemoprotection / Detox II TT TT<br />
rs1208 rs1208 NAT2 Chemoprotection / Detox II GG GG<br />
rs1799930 rs1799930 NAT2 Chemoprotection / Detox II AA GG<br />
rs1801279 rs1801279 NAT2 Chemoprotection / Detox II AA GG<br />
rs1041983 rs1041983 NAT2 Chemoprotection / Detox II TT CC<br />
rs35652124 rs35652124 NFE2L2 or NRF2 Chemoprotection / Detox II TT TC<br />
rs1962142 rs1962142 NFE2L2 or NRF2 Chemoprotection / Detox II AA GG<br />
rs6726395 rs6726395 NFE2L2 or NRF2 Chemoprotection / Detox II AA AG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs887829 rs887829 UGT1A1 Chemoprotection / Detox II TT CC<br />
rs3755319 rs3755319 UGT1A1 Chemoprotection / Detox II AA AC<br />
rs6742078 rs6742078 UGT1A1 G179250T Chemoprotection / Detox II TT GG<br />
rs4148323 rs4148323 UGT1A1 G211A Chemoprotection / Detox II AA GG<br />
rs1523127 rs1523127 PXR Chemoprotection / Detox II & III CC AA<br />
rs2062541 rs2062541 ABCC1 / MRP1 Chemoprotection / Detox III AA GG<br />
rs4148382 rs4148382 ABCC1 / MRP1 Chemoprotection / Detox III AA GG<br />
rs246240 rs246240 ABCC1 / MRP1 Chemoprotection / Detox III GG AG<br />
rs2273697 rs2273697 ABCC2 Chemoprotection / Detox III AA GG<br />
rs8187710 rs8187710 ABCC2 Chemoprotection / Detox III AA GG<br />
rs717620 rs717620 ABCC2 Chemoprotection / Detox III TT CC<br />
rs1405655 rs1405655 LXR Chemoprotection / Detox III TT TT<br />
rs17373080 rs17373080 LXR Chemoprotection / Detox III GG CC<br />
rs2695121 rs2695121 LXR Chemoprotection / Detox III TT CC<br />
rs3741049 rs3741049 ACAT1 G22670A Chemoprotection / Detox III / Bile Flow AA GG<br />
rs3733890 rs3733890 BHMT Chemoprotection / Detox III / Bile Flow AA GG<br />
rs7946 rs7946 PEMT Chemoprotection / Detox III / Bile Flow TT TT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs4244593 rs4244593 PEMT Chemoprotection / Detox III / Bile Flow TT GG<br />
rs7529229 rs7529229 IL6R Chemoprotection / Immune Factor CC TC<br />
rs7946 rs7946 PEMT Chemoprotection / Membranes health TT TT<br />
rs4244593 rs4244593 PEMT Chemoprotection / Membranes health TT GG<br />
rs429358 rs429358 APOE Chemoprotection / Oxidative Stress CC TT<br />
rs7412 rs7412 APOE Chemoprotection / Oxidative Stress CC CC<br />
rs769217 rs769217 CAT Chemoprotection / Oxidative Stress TT CT<br />
rs2300181 rs2300181 CAT C21068T Chemoprotection / Oxidative Stress TT CC<br />
rs4626565<br />
Last enzyme in Electron Transport<br />
Chain. ROS (Reactive Oxygen<br />
COX6C Chemoprotection / Oxidative Stress CC TT<br />
Species) scavenging.<br />
rs1050829 rs1050829 G6PD Chemoprotection / Oxidative Stress CC TT<br />
rs5030868 rs5030868 G6PD Chemoprotection / Oxidative Stress AA GG<br />
rs5030868 rs5030868 G6PD Chemoprotection / Oxidative Stress TT GG<br />
rs1050828 rs1050828 G6PD Chemoprotection / Oxidative Stress AA CC<br />
rs1050828 rs1050828 G6PD Chemoprotection / Oxidative Stress TT CC<br />
rs1050450 rs1050450 GPX1 Chemoprotection / Oxidative Stress AA GA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs2551715 Oxidised Glutathione reduction GSR Chemoprotection / Oxidative Stress TT CC<br />
rs1799836 rs1799836 MAOB Chemoprotection / Oxidative Stress TT TC<br />
rs35652124 rs35652124 NFE2L2 or NRF2 Chemoprotection / Oxidative Stress TT TC<br />
rs1962142 rs1962142 NFE2L2 or NRF2 Chemoprotection / Oxidative Stress AA GG<br />
rs6726395 rs6726395 NFE2L2 or NRF2 Chemoprotection / Oxidative Stress AA AG<br />
rs1800566 rs1800566 NQO1 Chemoprotection / Oxidative Stress AA GG<br />
rs854571 rs854571 PON1 Chemoprotection / Oxidative Stress TT TC<br />
rs662 rs662 PON1 Q192R Chemoprotection / Oxidative Stress TT CC<br />
rs4880 rs4880 SOD2 Chemoprotection / Oxidative Stress GG GG<br />
rs2069840 rs2069840 IL6<br />
Chemoprotection / Oxidative Stress /<br />
Immune Factor<br />
GG<br />
CC<br />
rs9282861 rs9282861 SULT1A1 Chemoprotection / Sulfonation CC CC<br />
rs182420 rs182420 SULT2A1 Chemoprotection / Sulfonation CC CT<br />
rs2547231 rs2547231 SULT2A1 Chemoprotection / Sulfonation AA CA<br />
rs11605924<br />
A (major) = Reduction in energy<br />
utilisation in a high fat diet.<br />
CRY2 Diet & Nutrition AA AA<br />
rs602662 rs602662 FUT2 Diet & Nutrition / B12 Status AA GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1801198 rs1801198 TCN2 Diet & Nutrition / B12 transport GG GC<br />
rs9606756 rs9606756 TCN2 Diet & Nutrition / B12 transport GG AA<br />
rs4148211<br />
rs708272<br />
The G (minor) allele is associated<br />
with: Increased total cholesterol<br />
levels Increased levels of LDL<br />
cholesterol but Decreased risk of<br />
developing type 2 diabetesThe A<br />
(major) allele is associated with:<br />
Increased levels of triglycerides<br />
(AA).Low levels of LDL cholesterol<br />
(AA).but Increased risk of<br />
developing type 2 diabetes (AA).<br />
The “C” (major) allele is associated<br />
with decreased levels of HDL (good<br />
cholesterol).The “A” allele is<br />
associated with increased HDL<br />
levels.<br />
ABCG8 Diet & Nutrition / Cholesterol Status GG AA<br />
CETP Diet & Nutrition / Cholesterol Status GG GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs3213445<br />
The Minor "C" allele is associated<br />
with:Carriers of the rare allele had<br />
significantly higher glutamyl<br />
transpeptidase (GGT), glutamic<br />
oxaloacetic transaminase (GOT) and<br />
glutamic pyruvate transaminase<br />
(GPT) activities.A higher fatty liver<br />
index.Triglycerides and fasting<br />
CPT1A Diet & Nutrition / Cholesterol Status CC TT<br />
glucose were significantly higher in<br />
C allele carriers.Insulin sensitivity<br />
was lower in C allele carriers.Total<br />
cholesterol and LDL-cholesterol<br />
tended to be higher in C allele<br />
carriers.<br />
rs174548 Diet & Nutrition / Cholesterol Status FADS1 Diet & Nutrition / Cholesterol Status GG CG<br />
rs482548<br />
(TT) Lower than common level of<br />
FADS enzyme activity.<br />
FADS2 Diet & Nutrition / Cholesterol Status TT CT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1535<br />
The Minor “G” allele is associated<br />
with:Decrease of Docosahexaenoic<br />
acid (DHA) (GG) . DHA is a type of<br />
omega-3, which is the primary<br />
component of a brain cell. The<br />
decreased ability transform fatty<br />
acids into Docosahexaenoic acid<br />
(DHA) results in a lower IQ for nonbreast<br />
fed children. Breast milk<br />
contains usable DHA.Some studies<br />
have shown that breastfed children<br />
with the GG genotype had the same<br />
IQs as the AG genotype. Both<br />
resulting in an average IQ in low the<br />
100s. While Non-breastfed children<br />
had IQs in the upper 90s.Low levels<br />
of High-density Lipoprotein<br />
Cholesterol (HDL-C) Moderately<br />
increases the risk for Type-2<br />
Diabetes.Intensive lifestyle<br />
FADS2 Diet & Nutrition / Cholesterol Status GG AG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs4846914<br />
The A (minor) allele is associated<br />
with increased levels of HDL<br />
cholesterol (good cholesterol) but<br />
GALNT2 Diet & Nutrition / Cholesterol Status GG GA<br />
also is associated with higher risk of<br />
hypertriglyceridemia.<br />
rs1800961 rs1800961 HNF4A Diet & Nutrition / Cholesterol Status TT CC<br />
rs1884614 rs1884614 HNF4A Diet & Nutrition / Cholesterol Status TT CC<br />
rs17700633<br />
The Minor "A" allele is associated<br />
with decreased HDL cholesterol .<br />
The G (major) allele is associated<br />
with increased HDL cholesterol.<br />
MC4R Diet & Nutrition / Cholesterol Status AA GA<br />
rs2229616<br />
The C (major) allele is associated<br />
with decreased HDL cholesterol<br />
levels.<br />
MC4R Diet & Nutrition / Cholesterol Status CC CC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs174537<br />
The G (major) allele is associated<br />
with increased LDL cholesterol (bad<br />
cholesterol) levels.The T (minor)<br />
allele is associated with decreased<br />
LDL cholesterol (bad cholesterol)<br />
levels.<br />
MYRF Diet & Nutrition / Cholesterol Status GG GT<br />
rs7946 rs7946 PEMT Diet & Nutrition / Cholesterol Status TT TT<br />
rs2016520<br />
The T (major) allele is associated<br />
with increased cholesterol levels.<br />
PPARD Diet & Nutrition / Cholesterol Status TT TT<br />
rs8192678 rs8192678 PPARGC1A Diet & Nutrition / Cholesterol Status TT CT<br />
rs646776<br />
The C (minor) allele is associated<br />
with increased levels of LDL (bad<br />
cholesterol).<br />
PSRC1 Diet & Nutrition / Cholesterol Status CC TT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs659366<br />
The "T" (minor) allele is associated<br />
with:More UCP2 production and<br />
higher levels of UCP2 protein<br />
Increased levels of HDL-Cholestrol<br />
and decreased levels of LDL-<br />
Cholestrol in a Chinese population.<br />
Reduced LDL particle size levels in a<br />
Chinese population.Increased<br />
energy expenditure.Decreased lipid<br />
oxidation Decreased high-sensitivity<br />
Cholesterol reactive protein (hs-<br />
CRP), a biomarker of inflammation.<br />
This decrease may indicate a lower<br />
level of inflammation in the body.<br />
More difficulty in losing weight<br />
under a very low-calorie program in<br />
Korean women.Higher fasting<br />
glucose levels in the blood.Higher<br />
risk of Diabetes (Type 2).Increased<br />
hardening of the arteries in women.<br />
UCP2 Diet & Nutrition / Cholesterol Status CC CT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs2073658<br />
The T (minor) allele is associated<br />
with increased LDL cholesterol.<br />
USF1 Diet & Nutrition / Cholesterol Status TT CC<br />
rs3733890 rs3733890 BHMT Diet & Nutrition / Choline Status GG GG<br />
rs2236225<br />
G = Decreased risk of choline<br />
deficiency.<br />
MTHFD1 Diet & Nutrition / Choline Status AA GA<br />
rs1805087 rs1805087 MTR Diet & Nutrition / Choline Status GG AA<br />
rs738409<br />
C = Decreased need for choline in<br />
the body.<br />
PNPLA3 Diet & Nutrition / Choline Status GG CC<br />
rs738409<br />
T (minor) = higher risk of fatty liver -<br />
encourage more Choline.Whites<br />
with fatty liver - which may be due<br />
to low choline - were more likely to<br />
carry the T allele.<br />
PNPLA3 Diet & Nutrition / Choline Status TT CC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs662799<br />
The G (minor) presumably reduces<br />
levels of APOA5 protein.<br />
Associations of APOA5 transcripts<br />
with PPARA and CPT1A1 transcripts<br />
suggest that APOA5 expression is<br />
intimately linked to hepatic lipid<br />
metabolism.<br />
The G (minor) allele is associated<br />
with:<br />
Each G=higher tryglycerides<br />
AG causes 1.4x higher early heart<br />
attack risk<br />
AG causes less weight gain on high<br />
fat diets<br />
GG causes 2x higher early heart<br />
attack risk<br />
GG causes less weight gain on high<br />
fat diets<br />
APOA5 Diet & Nutrition / Fat Breakdown GG AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1800562 rs1800562 HFE Diet & Nutrition / Iron Status AA GG<br />
rs1800562 rs1800562 HFE Diet & Nutrition / Iron Status AA GG<br />
rs1799945 rs1799945 HFE Diet & Nutrition / Iron Status GG CG<br />
rs6918586 rs6918586 HFE Diet & Nutrition / Iron Status TT CC<br />
rs3811647<br />
A = Increased Iron levels in the<br />
blood.<br />
TF Diet & Nutrition / Iron Status AA GG<br />
rs855791 A = Increased iron levels in blood. TMPRSS6 Diet & Nutrition / Iron Status AA GG<br />
rs4988235<br />
MCM6 has influence on the lactase<br />
LCT gene, rs182549 and is one of<br />
two SNPs that is associated with the<br />
primary haplotype associated with<br />
hypolactasia, more commonly<br />
known as lactose intolerance in<br />
European Caucasian populations.<br />
AA = Can digest milk .AG = Likely to<br />
be able to digest milk as an adult.<br />
GG = Likely lactose intolerant.<br />
MCM6 Diet & Nutrition / Lactose Tolerance GG AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs17300539<br />
rs174537<br />
The G (major) allele is associated<br />
with:Worse response to<br />
monounsaturated fat intake.GG =<br />
Higher BMI with increased<br />
monounsaturated fat intake (avoid<br />
monounsaturated fats)<br />
The G (major) allele is associated<br />
with:Possible decreased need for<br />
fish oil supplements to maintain<br />
healthy omega-3 fatty acid<br />
levelsIncreased levels of arachidonic<br />
acid in the body [R]. Arachidonic<br />
acid is an Omega-6 polyunsaturated<br />
fatty acid.TT = Possible increased<br />
need for fish oil supplements to<br />
maintain healthy omega-3 fatty acid<br />
levels.TT = Lower levels of<br />
arachidonic acid in the body.<br />
Arachidonic acid is an Omega-6<br />
polyunsaturated fatty acid.<br />
ADIPOQ Diet & Nutrition / Monounsaturated Fat GG GG<br />
MYRF Diet & Nutrition / Omega fatty acids TT GT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs894160<br />
rs1799986<br />
CC (major) =does worse with a high<br />
complex carb diet with regard to<br />
weight.People with the "T" or minor<br />
allele do better with complex carbs<br />
and worse with saturated fat. The T<br />
allele helps protects from weight<br />
gain from a higher complex carb<br />
diet, but causes weight gain on a<br />
lower carb complex carb diet and<br />
insulin resistance on a higher<br />
saturated fat diet.<br />
The C (major) does better with<br />
saturated fat (decreased weight<br />
gain)The AG genotype is associated<br />
with normal saturated fat levels.<br />
PLIN1<br />
Diet & Nutrition / Response to<br />
Carbohydrates<br />
LRP1 Diet & Nutrition / Saturated Fat TT CC<br />
rs744166 rs744166 STAT3 Diet & Nutrition / Saturated Fat GG AA<br />
rs8069645 rs8069645 STAT3 Diet & Nutrition / Saturated Fat GG AA<br />
rs651821<br />
TT = Decreased triglyceride levels.<br />
CC = Higher triglycerides.<br />
APOA5 Diet & Nutrition / Triglyceride Status CC TT<br />
CC<br />
CC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs3135506<br />
rs2072560<br />
rs2075291<br />
GG = Decreased triglyceride levels.<br />
CC = 34% Increase in triglyceride<br />
concentration.<br />
C = Lower levels of triglycerides.T =<br />
Higher levels of triglycerides.<br />
CC = Decreased risk of high<br />
triglyceride levels.AA = Greater than<br />
4x the risk for high triglyceride<br />
levels (n=Asian populations) AC = 4x<br />
higher risk for high triglyceride<br />
levels (n=Asian populations).<br />
APOA5 Diet & Nutrition / Triglyceride Status CC GG<br />
APOA5 Diet & Nutrition / Triglyceride Status TT CC<br />
APOA5 Diet & Nutrition / Triglyceride Status AA CC<br />
rs2266788 A = Decreased triglyceride levels. APOA5 Diet & Nutrition / Triglyceride Status GG AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs662799<br />
The G (minor) presumably reduces<br />
levels of APOA5 protein.<br />
Associations of APOA5 transcripts<br />
with PPARA and CPT1A1 transcripts<br />
suggest that APOA5 expression is<br />
intimately linked to hepatic lipid<br />
metabolism.<br />
The G (minor) allele is associated<br />
with:<br />
Each G=higher tryglycerides<br />
AG causes 1.4x higher early heart<br />
attack risk<br />
AG causes less weight gain on high<br />
fat diets<br />
GG causes 2x higher early heart<br />
attack risk<br />
GG causes less weight gain on high<br />
fat diets<br />
APOA5 Diet & Nutrition / Triglyceride Status GG AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs3213445<br />
rs174570<br />
rs13223993<br />
T = Decreased triglyceride levels.<br />
Triglycerides and fasting glucose<br />
were significantly higher in C allele<br />
carriers.<br />
CC = Normal levels of triglycerides.<br />
TT = Increased triglycerides.CT =<br />
Increased triglycerides.<br />
A = Increased triglyceride levelsG =<br />
Decreased triglycerides and<br />
increased HDL, lowering the risk of<br />
cardiovascular disease and obesity.<br />
CPT1A Diet & Nutrition / Triglyceride Status CC TT<br />
FADS2 Diet & Nutrition / Triglyceride Status TT CC<br />
IGFBP3 Diet & Nutrition / Triglyceride Status AA GA<br />
rs4889294 rs4889294 BCMO1 Diet & Nutrition / Vitamin A metabolism CC TT<br />
rs6564851 rs6564851 BCMO1 Diet & Nutrition / Vitamin A metabolism GG GG<br />
rs7501331 rs7501331 BCMO1 Diet & Nutrition / Vitamin A metabolism TT CC<br />
rs6420424 rs6420424 BCMO1 Diet & Nutrition / Vitamin A metabolism GG AA<br />
rs492602 rs492602 FUT2 Diet & Nutrition / Vitamin B 12 Status AA AA<br />
rs3741049 rs3741049 ACAT1 G22670A <strong>Gene</strong>ral Health / Blood Glucose AA GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs266729<br />
rs10885122<br />
Adiponectin is released in the<br />
bloodstream from fat cells, where it<br />
is involved in the control of fat<br />
metabolism and regulating glucose<br />
levels .CG was associated with type<br />
2 diabetes in the obese group only<br />
OR=2.45....P=0.02, and 59% risk of<br />
diabetes could be attributed to that.<br />
T=lower fasting glucoseG=higher<br />
fasting glucose<br />
ADIPOQ <strong>Gene</strong>ral Health / Blood Glucose GG CG<br />
ADRA2A <strong>Gene</strong>ral Health / Blood Glucose GG GG<br />
rs1405655 rs1405655 LXR <strong>Gene</strong>ral Health / Blood Glucose TT TT<br />
rs17373080 rs17373080 LXR <strong>Gene</strong>ral Health / Blood Glucose GG CC<br />
rs2695121 rs2695121 LXR <strong>Gene</strong>ral Health / Blood Glucose TT CC<br />
rs3135506<br />
The C (minor) allele is associated<br />
with:<br />
Higher risk of Cardiovascular<br />
disease.<br />
APOA5 <strong>Gene</strong>ral Health / Cardiovascular Health CC GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs662799<br />
The G (minor) presumably reduces<br />
levels of APOA5 protein.<br />
Associations of APOA5 transcripts<br />
with PPARA and CPT1A1 transcripts<br />
suggest that APOA5 expression is<br />
intimately linked to hepatic lipid<br />
metabolism.<br />
The G (minor) allele is associated<br />
with:<br />
Each G=higher tryglycerides<br />
AG causes 1.4x higher early heart<br />
attack risk<br />
AG causes less weight gain on high<br />
fat diets<br />
GG causes 2x higher early heart<br />
attack risk<br />
GG causes less weight gain on high<br />
fat diets<br />
APOA5 <strong>Gene</strong>ral Health / Cardiovascular Health GG AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1799945 rs1799945 HFE <strong>Gene</strong>ral Health / Cardiovascular Health GG CG<br />
rs3869109 rs3869109 HLA-C <strong>Gene</strong>ral Health / Cardiovascular Health GG AG<br />
rs5443 rs5443 HTR2A <strong>Gene</strong>ral Health / Cardiovascular Health TT CT<br />
rs8192678 rs8192678 PPARGC1A <strong>Gene</strong>ral Health / Cardiovascular Health TT CT<br />
rs4961<br />
The risk (T) allele responded better<br />
to diuretics and sodium-restricted<br />
diets, in that they tended to lower<br />
their blood pressure by ~10 mmHg<br />
points compared to rs4961(G;G)<br />
homozygotes similarly treated.<br />
ADD1 <strong>Gene</strong>ral Health / Clotting Factors TT GG<br />
rs1800775<br />
CETP encodes a protein that is<br />
involved in the transfer of<br />
cholesteryl ester from high density<br />
CETP <strong>Gene</strong>ral Health / Clotting Factors CC CC<br />
lipoprotein (HDL) to other<br />
lipoproteins.<br />
rs13146272 rs13146272 CYP4V2 <strong>Gene</strong>ral Health / Clotting Factors CC AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs3211719<br />
rs6025<br />
rs6048<br />
This gene encodes a protein called<br />
coagulation factor X that helps<br />
blood clot after an injury. Mutations<br />
can cause deficiency of the protein,<br />
which leads to excessive bleeding.<br />
he F5 gene encodes a protein that is<br />
called coagulation factor V. They<br />
that form blood clots to stop<br />
bleeding and trigger blood vessel<br />
repair after an injury. Mutations can<br />
cause abnormal bleeding.<br />
This gene encodes a protein called<br />
coagulation factor IX, which helps<br />
form blood clots. Mutations can<br />
cause blood clots to be unable to<br />
form and lead to excessive bleeding<br />
(hemophilia) or warfarin sensitivity,<br />
which can cause severe bleeding<br />
problems.<br />
F10 <strong>Gene</strong>ral Health / Clotting Factors GG AA<br />
F5 <strong>Gene</strong>ral Health / Clotting Factors TT CC<br />
F9 <strong>Gene</strong>ral Health / Clotting Factors GG GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1613662<br />
The gene encodes a protein that is a<br />
receptor for collagen and plays an<br />
important role in collagen-induced<br />
platelet aggregation and thrombus<br />
formation. Mutations of this gene<br />
can cause a bleeding disorder.<br />
GP6 Pro219Ser <strong>Gene</strong>ral Health / Clotting Factors GG AA<br />
rs9898 rs9898 HRG Pro204Ser <strong>Gene</strong>ral Health / Clotting Factors TT CT<br />
rs5918<br />
C=increased resistance to blood<br />
thinning side effect of aspirin.<br />
ITGB3 <strong>Gene</strong>ral Health / Clotting Factors CC TT<br />
rs2227589<br />
<strong>Gene</strong> variants associated with deep<br />
vein thrombosis.<br />
SERPINC1 <strong>Gene</strong>ral Health / Clotting Factors TT CC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs12273363<br />
rs6265<br />
Neurotrophins are chemicals that<br />
help to stimulate and control<br />
neurogenesis, BDNF being one of<br />
the most active. The C allele is<br />
associated with more with mood<br />
disorders than T.The minor C allele<br />
might increased activity in the<br />
amygdala (a region in brain). This<br />
might increase anxiety and<br />
emotional memory.<br />
Neurotrophins are chemicals that<br />
help to stimulate and control<br />
neurogenesis, BDNF being one of<br />
the most active. “T” allele carriers<br />
had lower blood BDNF than CC<br />
BDNF <strong>Gene</strong>ral Health / Cognitive CC TT<br />
BDNF <strong>Gene</strong>ral Health / Cognitive TT CT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs3738401<br />
rs10174400<br />
This gene encodes a protein that<br />
helps neurons grow and assist<br />
cortical development through its<br />
interaction with other proteins.<br />
Mutations are associated with<br />
higher risk for schizophrenia.<br />
Contributes to the generation and<br />
propagation of action potentials<br />
throughout the brain.This gene<br />
increases the capacity for<br />
information processing between the<br />
dorsolateral prefrontal cortex and<br />
dorsal anterior cingulate cortex,<br />
which support higher cognitive<br />
functions. T (minor allele)<br />
=significantly higher general<br />
cognitive or “G” independent of<br />
gender and age.<br />
DISC1 <strong>Gene</strong>ral Health / Cognitive AA GA<br />
SCN2A <strong>Gene</strong>ral Health / Cognitive CC CC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs4833095<br />
Toll Like Receptors get activated<br />
mainly by bacterial, viral and fungal<br />
products. Some of our junk protein<br />
can also activate TLRs. Once<br />
activated, they start a cascade that<br />
will produce cytokines and other<br />
inflammatory mediators to combat<br />
the infection.<br />
TLR1 <strong>Gene</strong>ral Health / Hayfever TT TT<br />
rs1049793 rs1049793 AOC1 / DAO <strong>Gene</strong>ral Health / Histamine Metabolism GG CG<br />
rs3918347 rs3918347 DAO <strong>Gene</strong>ral Health / Histamine Metabolism GG AG<br />
rs2073440 Histidine to Histamine HDC <strong>Gene</strong>ral Health / Histamine Metabolism GG TG<br />
rs901865 Histamine Receptor HRH1 <strong>Gene</strong>ral Health / Histamine Metabolism TT TC<br />
rs16940765 Histamine Receptor HRH4 <strong>Gene</strong>ral Health / Histamine Metabolism CC TT<br />
rs11662595 Histamine Receptor HRH4 <strong>Gene</strong>ral Health / Histamine Metabolism GG AA<br />
rs1799836 rs1799836 MAOB <strong>Gene</strong>ral Health / Histamine Metabolism TT TC<br />
rs1799929 rs1799929 NAT2 <strong>Gene</strong>ral Health / Histamine Metabolism TT TT<br />
rs1208 rs1208 NAT2 <strong>Gene</strong>ral Health / Histamine Metabolism GG GG<br />
rs1799930 rs1799930 NAT2 <strong>Gene</strong>ral Health / Histamine Metabolism AA GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1801279 rs1801279 NAT2 <strong>Gene</strong>ral Health / Histamine Metabolism AA GG<br />
rs1041983 rs1041983 NAT2 <strong>Gene</strong>ral Health / Histamine Metabolism TT CC<br />
rs9275224<br />
The G allele of rs9275224 is<br />
reported to be associated with Iga<br />
HLA-DQA2 <strong>Gene</strong>ral Health / Kidney Health GG GG<br />
Glomerulonephritis<br />
rs2007044 rs2007044 CACNA1C <strong>Gene</strong>ral Health / Mental Health GG AG<br />
rs16944 rs16944 IL1B <strong>Gene</strong>ral Health / Mental Health GG AA<br />
rs20541 rs20541 IL13 <strong>Gene</strong>ral Health / Skin Health GG GG<br />
rs10484554 rs10484554 HLA-C<br />
<strong>Gene</strong>ral Health / Skin Health / Psoriasis<br />
TT<br />
CC<br />
rs10885122<br />
These receptors have a critical role<br />
in regulating neurotransmitter<br />
release from your fight or flight<br />
system (sympathetic nerves and<br />
adrenergic neurons) in the brain.<br />
When activated, these receptors<br />
induce low heart rate, low blood<br />
pressure and sedation<br />
ADRA2A <strong>Gene</strong>ral Health / Stress Response TT GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1978340 rs1978340 GAD1 <strong>Gene</strong>ral Health / Stress Response AA GG<br />
rs3791878 rs3791878 GAD1 <strong>Gene</strong>ral Health / Stress Response GG GG<br />
rs3749034 rs3749034 GAD1 <strong>Gene</strong>ral Health / Stress Response GG GA<br />
rs6313 rs6313 HTR2A <strong>Gene</strong>ral Health / Stress Response AA GG<br />
rs6314 rs6314 HTR2A <strong>Gene</strong>ral Health / Stress Response AA GG<br />
rs5443 rs5443 HTR2A <strong>Gene</strong>ral Health / Stress Response TT CT<br />
rs6311 rs6311 HTR2A <strong>Gene</strong>ral Health / Stress Response TT CC<br />
rs2268494 rs2268494 OXTR <strong>Gene</strong>ral Health / Stress Response AA TT<br />
rs13316193 rs13316193 OXTR <strong>Gene</strong>ral Health / Stress Response TT TC<br />
rs1042778 rs1042778 OXTR <strong>Gene</strong>ral Health / Stress Response TT GT<br />
rs2268491 rs2268491 OXTR <strong>Gene</strong>ral Health / Stress Response TT CC<br />
rs53576 rs53576 OXTR (MAIN) <strong>Gene</strong>ral Health / Stress Response AA AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1867277<br />
The FOXE1 protein regulates the<br />
transcription of thyroglobulin (Tg)<br />
and thyroid peroxidase (TPO).AA=<br />
production of FOXE1 gene is<br />
increased (by forming a complex<br />
with USF1/USF2 transcription<br />
factors).The higher level of<br />
production of FOXE1 gene<br />
associated with development of<br />
thyroid cancer.<br />
FOXE1 <strong>Gene</strong>ral Health / Thyroid Health AA AA<br />
rs8192678 rs8192678 PPARGC1A <strong>Gene</strong>ral Health <strong>Report</strong> / Blood Glucose TT CT<br />
rs1800961 rs1800961 HNF4A GENERAL INFLAMMATION TT CC<br />
rs3761847 rs3761847 TRAF1 <strong>Gene</strong>ral Inflammation / Bone Health GG AA<br />
rs7041 rs7041 GC <strong>Gene</strong>ral Inflammation / Tuberculosis AA CC<br />
rs602662 rs602662 FUT2 GI Health / Crohn'S Disease AA GG<br />
rs2187668 rs2187668 HLA-DQA1 GI Health / Celiac & Gluten Intolerance TT CC<br />
rs7775228 rs7775228 HLA-DQB1 GI Health / Celiac & Gluten Intolerance CC TT<br />
rs1800871 rs1800871 IL10 GI Health / Celiac & Gluten Intolerance AG GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs6822844 rs6822844 IL2, IL21 GI Health / Celiac & Gluten Intolerance TT GG<br />
rs3741049 rs3741049 ACAT1 G22670A GI Health / Dysbiosis AA GG<br />
rs2069840 rs2069840 IL6 GI Health / Gastrointestinal Immunology GG CC<br />
rs601338 rs601338 FUT2 GI Health / H Antigen AA GG<br />
rs602662 rs602662 FUT2 GI Health / H Antigen AA GG<br />
rs1047781 rs1047781 FUT2 GI Health / H Antigen TT AA<br />
rs492602 rs492602 FUT2 GI Health / H Antigen GG AA<br />
rs1878672 rs1878672 IL10 GI Health / H.Pylori CC CC<br />
rs10156191 rs10156191 AOC1 / DAO GI Health / Histamine Intolerance TT CT<br />
rs1049793 rs1049793 AOC1 / DAO GI Health / Histamine Intolerance GG CG<br />
rs3918347 rs3918347 DAO GI Health / Histamine Intolerance GG AG<br />
rs602662 rs602662 FUT2 GI Health / IBS AA GG<br />
rs5443 rs5443 HTR2A GI Health / IBS TT CT<br />
rs6311 rs6311 HTR2A GI Health / IBS TT CC<br />
rs1800871 rs1800871 IL10 GI Health / IBS AA GG<br />
rs10210302 rs10210302 ATG16L1 GI Health / IgA / Crohn's disease TT TT<br />
rs1061622 rs1061622 TNFRSF1B GI Health / Inflammation / IBS GG TG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs854571 rs854571 PON1 GI Health / Microbial defence / IBD TT TC<br />
rs662 rs662 PON1 Q192R GI Health / Microbial defence / IBD TT CC<br />
rs7946 rs7946 PEMT GI Health / Permeability TT TT<br />
rs4244593 rs4244593 PEMT GI Health /Permeability TT GG<br />
rs2289252<br />
This gene encodes a protein called<br />
coagulation factor XI that helps<br />
blood clot after an injury. Mutations<br />
F11,F11-AS1 Health <strong>Report</strong> / Clotting Factors TT CT<br />
can cause deficiency of the protein,<br />
which leads to excessive bleeding.<br />
rs6994992 rs6994992 NRG1 Health <strong>Report</strong> / Cognitive TT CT<br />
rs3746544 rs3746544 SNAP25 Health <strong>Report</strong> / Cognitive TT TT<br />
rs4343 rs4343 ACE G2328A Health <strong>Report</strong> / Kidney Health GG GA<br />
rs4343 rs4343 ACE G2328A Health <strong>Report</strong> / Stress Response GG GA<br />
rs429358 rs429358 APOE Immune Factors & Inflammation CC TT<br />
rs7412 rs7412 APOE Immune Factors & Inflammation CC CC<br />
rs10210302 rs10210302 ATG16L1 Immune Factors & Inflammation TT TT<br />
rs7216389 rs7216389 GSDMB Immune Factors & Inflammation TT CT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs6822844 rs6822844 IL2, IL21 Immune Factors & Inflammation TT GG<br />
rs2280714<br />
IFN-regulatory factor 5 gene<br />
increases IFN-alpha. Interferon<br />
alpha induces the kynurenine<br />
pathway and is associated with<br />
depression because of NMDA<br />
overactivity.The C (minor) allele is<br />
associated with:Possible increased<br />
risk of depression when paired with<br />
STAT4 T allele. Increased levels of<br />
IFN-alpha protein.<br />
IRF5 Immune Factors & Inflammation CC CT<br />
rs1405655 rs1405655 LXR Immune Factors & Inflammation TT TT<br />
rs17373080 rs17373080 LXR Immune Factors & Inflammation GG CC<br />
rs2695121 rs2695121 LXR Immune Factors & Inflammation TT CC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs4833095<br />
Toll Like Receptors get activated<br />
mainly by bacterial, viral and fungal<br />
products. Some of our junk protein<br />
can also activate TLRs. Once<br />
activated, they start a cascade that<br />
will produce cytokines and other<br />
inflammatory mediators to combat<br />
the infection.<br />
TLR1 Immune Factors & Inflammation TT TT<br />
rs3775291 rs3775291 TLR3 Immune Factors & Inflammation CC CC<br />
rs1061622 rs1061622 TNFRSF1B Immune Factors & Inflammation GG TG<br />
rs3761847 rs3761847 TRAF1 Immune Factors & Inflammation GG AA<br />
rs7775228 rs7775228 HLA-DQB1<br />
Immune Factors & Inflammation /<br />
Allergies<br />
CC<br />
TT<br />
rs231779 rs231779 CTLA4<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
TT<br />
TT<br />
rs231775 rs231775 CTLA4<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
GG<br />
GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs3087243 rs3087243 CTLA4<br />
rs11575839 rs11575839 HLA-C<br />
rs10484554 rs10484554 HLA-C<br />
rs2187668 rs2187668 HLA-DQA1<br />
rs9275224<br />
rs3135391<br />
rs3135388<br />
Helps to recognise foreign particles<br />
when they enter our body and fight<br />
them away in an immune response.<br />
Increases risk for<br />
autoimmunity/inflammation to<br />
toxins or infections. Each A<br />
allele=very significant increase in<br />
autoimmune disease risk.<br />
rs3135388<br />
HLA-DQA2<br />
HLA-DRB1<br />
HLA-DRB5/HLA-DRB1/HLA-DRA/HLA-<br />
DQB1/ HLA-DQA1<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
A,G<br />
AA<br />
TT<br />
AA<br />
AA<br />
AA<br />
AA<br />
GG<br />
GG<br />
CC<br />
CC<br />
GG<br />
GG<br />
GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs11889341 rs11889341 STAT4<br />
rs10181656 rs10181656 STAT4<br />
rs897200 rs897200 STAT4<br />
rs1517352 rs1517352 STAT4<br />
rs6677604<br />
rs1990760<br />
C3, a protein coding gene, is part of<br />
a system of proteins that fight<br />
against disease.<br />
This gene encodes a protein that is<br />
involved with RNA translation<br />
initiation, splicing, and assembly. It<br />
is also involved in the formation of<br />
embryos and sperm and cellular<br />
growth and division.<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
Immune Factors & Inflammation /<br />
Autoimmunity<br />
CFH Immune Factors & Inflammation / IgA AA GG<br />
IFIH1 Immune Factors & Inflammation / IgA TT CC<br />
TT<br />
GG<br />
AA<br />
CC<br />
TT<br />
GG<br />
CC<br />
AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs2229765<br />
IGF1R encodes for insulin growth<br />
factor receptors; overexpression<br />
can stop cell death by enhancing<br />
cell survival and is present in<br />
malignant tissues. AA= lower levels<br />
of IGF-1 protein.Increased longevity<br />
in males 15% of people have AA.<br />
Most people have AG.<br />
IGF1R Immune Factors & Inflammation / IgA AA GG<br />
rs9357155 rs9357155 PSMB8 Immune Factors & Inflammation / IgA AA GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs2814778<br />
rs2814778 is within the DARC gene,<br />
which encodes the Duffy blood<br />
group antigen [PMID 7663520]. This<br />
SNP shows an almost perfectly fixed<br />
difference in frequency between<br />
Europeans and those with African<br />
ancestry. (One exception appears to<br />
be a certain population of Czech<br />
gypsies, and certain non-Ashkenazi<br />
Jewish populations.) Additionally<br />
the Namibian San samples of the<br />
CEPH-HGDP are, uncharacteristically<br />
for Africans, all AA homozygotes for<br />
this SNP.The rs2814778 (G) allele is<br />
associated with African populations,<br />
while rs2814778 (A) is associated<br />
with European populations and<br />
southwestern Native American<br />
populations.<br />
ACKR1 Immune Factors & Inflammation / IgE CC TT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs2251746<br />
This gene encodes a protein that<br />
initiates the body's allergic<br />
response. High levels of serum IgE<br />
are associated with allergies, and<br />
are mediators of autoimmune<br />
diseases.<br />
FCER1A Immune Factors & Inflammation / IgE CC TT<br />
rs2858331 rs2858331 HLA-DQB1 Immune Factors & Inflammation / IgE GG AG<br />
rs2240032 rs2240032 RAD50 Immune Factors & Inflammation / IgE TT CC<br />
rs2040704 rs2040704 RAD50 Immune Factors & Inflammation / IgE GG AA<br />
rs1801274<br />
This gene encodes a protein that<br />
plays an important role in the<br />
immune system response by<br />
fighting off pathogens. It is<br />
FCGR2A Immune Factors & Inflammation / IgG GG GG<br />
associated with higher susceptibility<br />
to arthritis, lupus, and malaria.<br />
rs10757278 rs10757278 DMRTA1<br />
Immune Factors & Inflammation /<br />
Microbial defence<br />
GG<br />
GG<br />
rs854571 rs854571 PON1<br />
Immune Factors & Inflammation /<br />
Microbial defence<br />
TT<br />
TC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs662 rs662 PON1 Q192R<br />
rs1816702 rs1816702 TLR2<br />
rs4986791 rs4986791 TLR4<br />
rs4986790 rs4986790 TLR4<br />
rs6853 rs6853 MYD88<br />
rs4986790 rs4986790 TLR4<br />
Immune Factors & Inflammation /<br />
Microbial defence<br />
Immune Factors & Inflammation /<br />
Microbial defence<br />
Immune Factors & Inflammation /<br />
Microbial defence<br />
Immune Factors & Inflammation /<br />
Microbial defence<br />
Immune Factors & Inflammation /<br />
Vaccine Response<br />
Immune Factors & Inflammation /<br />
Vaccine Response<br />
rs1800871 rs1800871 IL10 Immune Factors and Inflammation AA GG<br />
rs1878672 rs1878672 IL10 Immune Factors and Inflammation CC CC<br />
rs3024490 rs3024490 IL10 Immune Factors and Inflammation AA CC<br />
rs20541 rs20541 IL13 Immune Factors and Inflammation AA GG<br />
rs1143643 rs1143643 IL1B Immune Factors and Inflammation CC CC<br />
rs16944 rs16944 IL1B Immune Factors and Inflammation GG AA<br />
TT<br />
TT<br />
TT<br />
GG<br />
GG<br />
GG<br />
CC<br />
CC<br />
CC<br />
AA<br />
AA<br />
AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1801275 rs1801275 IL4R Immune Factors and Inflammation GG AG<br />
rs2069812 Anti-inflamatory cytokines. IL5 Immune Factors and Inflammation GG AG<br />
rs2069840 rs2069840 IL6 Immune Factors and Inflammation GG CC<br />
rs7529229 rs7529229 IL6R Immune Factors and Inflammation CC TC<br />
rs6994992 rs6994992 NRG1 Immune Factors and Inflammation TT CT<br />
rs10757278 rs10757278 STAT1 Immune Factors and Inflammation GG GG<br />
rs6503691 rs6503691 STAT3 Immune Factors and Inflammation CC CC<br />
rs744166 rs744166 STAT3 Immune Factors and Inflammation AA AA<br />
rs8069645 rs8069645 STAT3 Immune Factors and Inflammation GG AA<br />
rs361525 Tumor Necrosis Factor TNF -238 Immune Factors and Inflammation AA GG<br />
rs1800629 Tumor Necrosis Factor TNF -308 Immune Factors and Inflammation AA GG<br />
rs2282679 rs2282679 GC<br />
Immune Modulation / Vitamin D &<br />
GcMAF<br />
GG<br />
TT<br />
rs7041 rs7041 GC<br />
Immune Modulation / Vitamin D &<br />
GcMAF<br />
CC<br />
CC<br />
rs1851024 rs1851024 GC<br />
Immune Modulation / Vitamin D &<br />
GcMAF<br />
GG<br />
AA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs703842 rs703842 CYP27B1<br />
rs10500804 rs10500804 CYP2R1<br />
rs12794714 rs12794714 CYP2R1<br />
rs2060793 rs2060793 CYP2R1<br />
rs61495246 rs61495246 CYP2R1<br />
rs1544410 rs1544410 VDR Bsm1<br />
rs731236 rs731236 VDR Taql<br />
Immune Modulation / Vitamin D<br />
Metabolism<br />
Immune Modulation / Vitamin D<br />
Metabolism<br />
Immune Modulation / Vitamin D<br />
Metabolism<br />
Immune Modulation / Vitamin D<br />
Metabolism<br />
Immune Modulation / Vitamin D<br />
Metabolism<br />
Immune Modulation / Vitamin D<br />
Metabolism<br />
Immune Modulation / Vitamin D<br />
Metabolism<br />
rs3733890 rs3733890 BHMT Methylation AA GG<br />
rs567754 rs567754 BHMT-02 Methylation TT TT<br />
rs2851391 Homocysteine to Cystathionine. CBS Methylation TT TC<br />
rs4920037 Homocysteine to Cystathionine. CBS Methylation AA GA<br />
AA<br />
GG<br />
AA<br />
AA<br />
GG<br />
TT<br />
GG<br />
AA<br />
TG<br />
GA<br />
AG<br />
AA<br />
TT<br />
GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs234706 Homocysteine to Cystathionine CBS C699T Methylation AA GA<br />
rs1677693 FOLR2 to DHF, DHF to THF DHFR Methylation TT GT<br />
rs705415 rs705415 DMGDH Methylation TT CT<br />
rs2071010<br />
Binds folates to transport the inside<br />
cells.<br />
FOLR1 Methylation AA GG<br />
rs651933<br />
Binds folates to transport the inside<br />
cells.<br />
FOLR2 Methylation GG AA<br />
rs17851582<br />
Arginine to Guanidoacetate to<br />
Creatine (SAMe)<br />
GAMT Methylation AA GG<br />
rs55776826<br />
Arginine to Guanidoacetate to<br />
Creatine (SAMe)<br />
GAMT Methylation TT CC<br />
rs3780127 rs3780127 GGH Methylation AA GG<br />
rs3780126 GGH Methylation TT GA<br />
rs72558181 rs72558181 MAT1A Methylation TT CC<br />
rs2236225<br />
THF to Purines (ATP & GTP) & 10-<br />
Formyl-THF to 5,10 Methenyl THF to<br />
MTHFD1 Methylation AA GA<br />
5,10 Methylene THF<br />
rs1801131 rs1801131 MTHFR A1298C Methylation GG TT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1801133 rs1801133 MTHFR C677T Methylation AA GG<br />
rs2733103<br />
Helps regulate carbon flow through<br />
the folate-dependent onecarbonmetabolic<br />
network that<br />
supplies carbon for the biosynthesis<br />
of purines,thymidine, and amino<br />
acids.<br />
MTHFS Methylation TT CC<br />
rs2275565 rs2275565 MTR Methylation TT GG<br />
rs3820571 rs3820571 MTR Methylation GG GG<br />
rs2275568 rs2275568 MTR Methylation TT CC<br />
rs10925257 rs10925257 MTR Methylation GG AA<br />
rs11799670 rs11799670 MTR Methylation GG AA<br />
rs1805087 rs1805087 MTR Methylation GG AA<br />
rs1532268 rs1532268 MTRR Methylation TT CC<br />
rs1802059 rs1802059 MTRR -11 A664A Methylation AA GG<br />
rs1801394 rs1801394 MTRR A66G Methylation GG GG<br />
rs162036 rs162036 MTRR K350A Methylation GG AG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs7946 rs7946 PEMT Methylation TT TT<br />
rs4244593 rs4244593 PEMT Methylation TT GG<br />
rs1051266 rs1051266 SLC19A1 Methylation CC CC<br />
rs2274894<br />
iNOS, a reactive free radical<br />
generator.<br />
NOS2 Mitochondrial Function TT TT<br />
rs2248814<br />
iNOS, a reactive free radical<br />
generator<br />
NOS2 Mitochondrial Function AA AA<br />
rs1800779 eNOS production. NOS3 Mitochondrial Function GG AA<br />
rs8192678 rs8192678 PPARGC1A Mitochondrial Function TT CT<br />
rs2970869 rs2970869 PPARGC1A Mitochondrial Function TT CC<br />
rs3774923 rs3774923 PPARGC1A Mitochondrial Function TT CC<br />
rs4880 rs4880 SOD2 Mitochondrial Function GG GG<br />
rs659366<br />
UCP are mitochondrial transporter<br />
proteins that create proton leaks<br />
across the inner mitochondrial<br />
membrane.<br />
UCP2 Mitochondrial Function CC CT<br />
rs7134594<br />
Conversion of vitamin B-12 into<br />
adenosylcobalamin<br />
MMAB<br />
Mitochondrial Function / Cobalamin<br />
Contribution<br />
CC<br />
CC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs4147730<br />
rs1142530<br />
rs2075626<br />
Electron Transport Chain, ATP<br />
Production<br />
Electron Transport Chain, ATP<br />
Production<br />
Electron Transport Chain, ATP<br />
Production<br />
NDUFS3 Mitochondrial Function / Complex I AA GA<br />
NDUFS7 Mitochondrial Function / Complex I TT TT<br />
NDUFS8 Mitochondrial Function / Complex I CC TT<br />
rs3741049 rs3741049 ACAT1 G22670A Mitochondrial Function / Krebs Cycle AA GG<br />
rs2302729 rs2302729 CACNA1C Neurotransmitters TT CC<br />
rs2007044 rs2007044 CACNA1C Neurotransmitters GG AG<br />
rs216013 rs216013 CACNA1C Neurotransmitters GG AA<br />
rs1006737 rs1006737 CACNA1C / SLC36A3 Neurotransmitters AA GG<br />
rs72547566 Adrenaline metabolism (NAD,B1) ALDH3A2<br />
Neurotransmitters / Adrenaline<br />
Metabolism<br />
TT<br />
CC<br />
rs10994336 rs10994336 ANK3 Neurotransmitters / Biopterin Cycle TT CC<br />
rs4948418 rs4948418 ANK3 Neurotransmitters / Biopterin Cycle TT CC<br />
rs9804190 rs9804190 ANK3 Neurotransmitters / Biopterin Cycle TT CC<br />
rs1677693 Biopterin Recycling DHFR Neurotransmitters / Biopterin Cycle TT GT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs3783642 rs3783642 GCH1 Neurotransmitters / Biopterin Cycle CC TC<br />
rs3783641 rs3783641 GCH1 Neurotransmitters / Biopterin Cycle AA TA<br />
rs3783637 rs3783637 GCH1 Neurotransmitters / Biopterin Cycle TT CC<br />
rs8007267 rs8007267 GCH1 C36378991T Neurotransmitters / Biopterin Cycle TT CT<br />
rs10483639 rs10483639 GCH1 G55306457C Neurotransmitters / Biopterin Cycle CC GC<br />
rs1800779 Biopterin Recycling. NOS3 Neurotransmitters / Biopterin Cycle GG AA<br />
rs5030858 rs5030858 PAH Neurotransmitters / Biopterin Cycle AA GG<br />
rs5030849 rs5030849 PAH Neurotransmitters / Biopterin Cycle TT CC<br />
rs6356 Tyrosine to L-Dopa TH Neurotransmitters / Biopterin Cycle TT CC<br />
rs2070762 Tyrosine to L-Dopa TH Neurotransmitters / Biopterin Cycle GG AG<br />
rs4633 rs4633 COMT H62H<br />
Neurotransmitters / Dopamine &<br />
Adrenaline Metabolism<br />
CC<br />
CC<br />
rs4680 rs4680 COMT V158M<br />
Neurotransmitters / Dopamine &<br />
Adrenaline Metabolism<br />
AA<br />
GG<br />
rs737866 rs737866 COMT/TXNRD2 A4251G<br />
Neurotransmitters / Dopamine &<br />
Adrenaline Metabolism<br />
TT<br />
TC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs2020917 rs2020917 COMT/TXNRD2 C4622T<br />
rs1611115 rs1611115 DBH<br />
rs1108580 rs1108580 DBH<br />
rs1018381<br />
Synaptic vesicle trafficking,<br />
neurotransmitter release.Role in<br />
regulation of cell surface exposure<br />
of Dopamine Receptor 2<br />
DTNBP1<br />
Neurotransmitters / Dopamine &<br />
Adrenaline Metabolism<br />
Neurotransmitters / Dopamine<br />
Metabolism<br />
Neurotransmitters / Dopamine<br />
Metabolism<br />
Neurotransmitters / Dopamine<br />
Metabolism<br />
rs686 rs686 DRD1 Neurotransmitters / Dopamine Receptor AA GA<br />
rs4532 rs4532 DRD1 Neurotransmitters / Dopamine Receptor CC CT<br />
rs265981 rs265981 DRD1 Neurotransmitters / Dopamine Receptor AA AG<br />
rs1079727 rs1079727 DRD2 Neurotransmitters / Dopamine Receptor CC TT<br />
rs1125394 rs1125394 DRD2 Neurotransmitters / Dopamine Receptor CC TT<br />
rs4436578 rs4436578 DRD2 Neurotransmitters / Dopamine Receptor CC TT<br />
rs1801028 rs1801028 DRD2 Neurotransmitters / Dopamine Receptor CG GG<br />
rs4648317 rs4648317 DRD2 Neurotransmitters / Dopamine Receptor AA GG<br />
CC<br />
TT<br />
GG<br />
AA<br />
CT<br />
CC<br />
AG<br />
GG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs4648319 rs4648319 DRD2 Neurotransmitters / Dopamine Receptor AA GG<br />
rs6279 rs6279 DRD2 Neurotransmitters / Dopamine Receptor GG GC<br />
rs1076560 rs1076560 DRD2 Neurotransmitters / Dopamine Receptor AA CC<br />
rs1079596 rs1079596 DRD2 Neurotransmitters / Dopamine Receptor TT CC<br />
rs1079597 rs1079597 DRD2 Neurotransmitters / Dopamine Receptor TT CC<br />
rs2283265 rs2283265 DRD2 Neurotransmitters / Dopamine Receptor AA CC<br />
rs1800498 rs1800498 DRD2 Neurotransmitters / Dopamine Receptor AA AA<br />
rs1800497 rs1800497 DRD2 (MAIN) Neurotransmitters / Dopamine Receptor AA GG<br />
rs6277 rs6277 DRD2 (MAIN) Neurotransmitters / Dopamine Receptor AA GA<br />
rs1799978 rs1799978 DRD2 A4651G Neurotransmitters / Dopamine Receptor CC TT<br />
rs2242592 rs2242592 DRD2 C71572T Neurotransmitters / Dopamine Receptor AA GA<br />
rs2242592 rs2242592 DRD2 C71572T Neurotransmitters / Dopamine Receptor GG GA<br />
rs12364283 rs12364283 DRD2 T4047C Neurotransmitters / Dopamine Receptor GG AA<br />
rs6280 rs6280 DRD3 Neurotransmitters / Dopamine Receptor CC TT<br />
rs167771 rs167771 DRD3 Neurotransmitters / Dopamine Receptor GG AA<br />
rs1049793 rs1049793 AOC1 / DAO Neurotransmitters / Dopamine Synthesis GG CG<br />
rs3918347 rs3918347 DAO Neurotransmitters / Dopamine Synthesis GG AG
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs3741775 rs3741775 DAO Neurotransmitters / Dopamine Synthesis CC AC<br />
rs6323 rs6323 MAO A R297R<br />
rs1137070 rs1137070 MAOA<br />
rs1799836 rs1799836 MAOB<br />
rs3219151<br />
GABA receptor. TT=higher cortisol,<br />
CT=Intermediate.<br />
Neurotransmitters / Dopamine,<br />
Serotonin, Adrenaline, Noradrenaline<br />
Metabolism<br />
Neurotransmitters / Dopamine,<br />
Serotonin, Adrenaline, Noradrenaline<br />
Metabolism<br />
Neurotransmitters / Dopamine,<br />
Serotonin, Adrenaline, Noradrenaline<br />
Metabolism<br />
GABRA6 Neurotransmitters / Glutamate ➜ GABA TT CT<br />
rs3791878 rs3791878 GAD1 Neurotransmitters / Glutamate ➜ GABA GG GG<br />
rs3749034 rs3749034 GAD1 Neurotransmitters / Glutamate ➜ GABA GG GA<br />
rs2241165 rs2241165 GAD1 Neurotransmitters / Glutamate ➜ GABA CC CT<br />
rs4668338 rs4668338 GAD1 Neurotransmitters / Glutamate ➜ GABA GG CC<br />
rs6994992 rs6994992 NRG1 Neurotransmitters / Glutamate ➜ GABA TT CT<br />
GG<br />
TT<br />
TT<br />
TT<br />
CC<br />
TC
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs671 rs671 ALDH2<br />
rs2238151 rs2238151 ALDH2<br />
rs40147<br />
rs3785143<br />
rs3785152<br />
rs2242446<br />
Noradrenaline Transporter,<br />
reuptake of noradrenaline<br />
Noradrenaline Transporter,<br />
reuptake of noradrenaline<br />
Noradrenaline Transporter,<br />
reuptake of noradrenaline<br />
Noradrenaline Transporter,<br />
reuptake of noradrenaline<br />
SLC6A2<br />
SLC6A2<br />
SLC6A2<br />
SLC6A2 C5884T<br />
rs2167364 rs2167364 DDC<br />
rs6592961 rs6592961 DDC<br />
rs2329340 rs2329340 DDC<br />
Neurotransmitters / Noradrenaline<br />
Metabolism<br />
Neurotransmitters / Noradrenaline<br />
Metabolism<br />
Neurotransmitters / Noradrenaline<br />
Metabolism<br />
Neurotransmitters / Noradrenaline<br />
Metabolism<br />
Neurotransmitters / Noradrenaline<br />
Metabolism<br />
Neurotransmitters / Noradrenaline<br />
Metabolism<br />
Neurotransmitters / Serotonin &<br />
Dopamine Synthesis<br />
Neurotransmitters / Serotonin &<br />
Dopamine Synthesis<br />
Neurotransmitters / Serotonin &<br />
Dopamine Synthesis<br />
AA<br />
TT<br />
AA<br />
TT<br />
TT<br />
CC<br />
CC<br />
AA<br />
TT<br />
GG<br />
CC<br />
GA<br />
CC<br />
CC<br />
TT<br />
TT<br />
GG<br />
CT
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs1451375 rs1451375 DDC<br />
Neurotransmitters / Serotonin &<br />
Dopamine Synthesis<br />
rs11077820 Serotonin to Melatonin AANAT<br />
Neurotransmitters / Serotonin ➜<br />
Melatonin<br />
CC<br />
TT<br />
rs7997012 rs7997012 HTR2A Neurotransmitters / Serotonin Receptors AA AG<br />
rs4565946 rs4565946 TPH2 Neurotransmitters I Biopterin I Serotonin GG TT<br />
rs4570625 rs4570625 TPH2 Neurotransmitters I Biopterin I Serotonin GG GG<br />
rs2851391<br />
It is responsible for using vitamin B6<br />
to convert amino acids called<br />
homocysteine and serine to a<br />
molecule called cytathionine.<br />
Another enzyme converts<br />
cystathionine to cysteine, which is<br />
used to build proteins or is broken<br />
down and excreted in urine.<br />
CBS Trans-sulfuration TT TC<br />
AA<br />
CA
Lifestyle Genomics Research Centre Ltd.<br />
Plymouth Science Park, 1 Davy Road, Derriford, Plymouth, PL6 8BX , UNITED KINGDOM<br />
contact us<br />
SAMPLE PROCESSED BY:<br />
METHOD:<br />
DATA CHECKED FOR ACCURACY BY:<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
Fluidigm TaqMan probe-based SNP genotyping / Illumina MiSeq, Next <strong>Gene</strong>ration<br />
sequencing<br />
Genome Centre at BARTS and The London School of Medicine and Dentistry<br />
NOTES:<br />
genotype has no risk alleles<br />
genotype has one risk allele<br />
READ THIS FIRST JOIN US ON FACEBOOK BOOK AN APPOINTMENT<br />
genotype has two risk allels<br />
either allele can be a risk or be<br />
beneficial<br />
Ctr+F to activate Search Engine: Look up genes, rsids, pathways / traits<br />
SNP RSID INTERPRET ☞ PARENT GENE & VARIANT ⚛ CATEGORY ☤ / PATHWAY ⥹ RISK ALLELES !! cp00035774<br />
rs4920037<br />
rs234706<br />
It is responsible for using vitamin B6<br />
to convert amino acids called<br />
homocysteine and serine to a<br />
molecule called cytathionine.<br />
Another enzyme converts<br />
cystathionine to cysteine, which is<br />
used to build proteins or is broken<br />
down and excreted in urine.<br />
AA=Increased responsiveness to<br />
homocysteine-lowering effects of<br />
folic acid. Limits homocysteine into<br />
downstream path. People with CBS<br />
mutations will need to be careful<br />
with sulphur containing<br />
supplements. Increased risk for<br />
ammonia detoxification issues.<br />
CBS Trans-sulfuration AA GA<br />
CBS C699T Trans-sulfuration AA GA<br />
rs2273684 rs2273684 GSS Trans-sulfuration TT GG<br />
rs6088659 rs6088659 GSS A5997G Trans-sulfuration TT CC