- Page 2:
The Staphylococcus aureus secretome
- Page 6:
RIJKSUNIVERSITEIT GRONINGEN The Sta
- Page 10:
Paranimfen: Thijs R.H.M. Kouwen Mon
- Page 14:
Table of contents Chapter 1. Genera
- Page 18:
Chapter 1 Introduction and scope of
- Page 22:
Introduction and scope of this thes
- Page 26:
Introduction and scope of this thes
- Page 30:
Chapter 2 Mapping the pathways to s
- Page 34:
Mapping the pathways to staphylococ
- Page 38:
Mapping the pathways to staphylococ
- Page 42:
Mapping the pathways to staphylococ
- Page 46:
Mapping the pathways to staphylococ
- Page 50:
Mapping the pathways to staphylococ
- Page 54:
Mapping the pathways to staphylococ
- Page 58:
Mapping the pathways to staphylococ
- Page 62:
Mapping the pathways to staphylococ
- Page 66:
Mapping the pathways to staphylococ
- Page 70:
Mapping the pathways to staphylococ
- Page 74:
Mapping the pathways to staphylococ
- Page 78:
Mapping the pathways to staphylococ
- Page 82:
Mapping the pathways to staphylococ
- Page 86:
Mapping the pathways to staphylococ
- Page 90:
Mapping the pathways to staphylococ
- Page 94:
Mapping the pathways to staphylococ
- Page 98:
Mapping the pathways to staphylococ
- Page 104:
“Music is the medicine of the min
- Page 108:
Chapter 3 Summary Sequencing of at
- Page 112:
Chapter 3 transcription analyses, S
- Page 116:
Chapter 3 Lina et al., 2003). The a
- Page 120:
Chapter 3 (Supplemental tables IVb)
- Page 124:
Chapter 3 Figure 1. Characterizatio
- Page 128:
Chapter 3 Figure 3. Relative amount
- Page 132:
Chapter 3 strain of a patient who s
- Page 136:
“Music is my religion” -Johnny
- Page 140:
Chapter 4 Summary Staphylococcus au
- Page 144:
Chapter 4 secretion pathways have b
- Page 148:
Chapter 4 has been shown that S. au
- Page 152:
Chapter 4 Table 2 Plasmids and bact
- Page 156:
Chapter 4 Table 3 Primers used in t
- Page 160:
Chapter 4 containing X-gal and incu
- Page 164:
Chapter 4 assay, 25 L4-stage nemato
- Page 168:
Chapter 4 D O 540 18 16 14 12 10 8
- Page 172:
Chapter 4 These findings suggested
- Page 176:
Chapter 4 precursors by lgt mutant
- Page 180:
Chapter 4 aureus lgt mutants may be
- Page 186:
Chapter 5 Synthetic effects of secG
- Page 190:
Synthetic effects of secG and secY2
- Page 194:
Synthetic effects of secG and secY2
- Page 198:
Synthetic effects of secG and secY2
- Page 202:
Synthetic effects of secG and secY2
- Page 206:
Synthetic effects of secG and secY2
- Page 210:
Synthetic effects of secG and secY2
- Page 214:
Synthetic effects of secG and secY2
- Page 218:
109
- Page 222:
Chapter 6 Partially overlapping sub
- Page 226:
Partially overlapping substrate spe
- Page 230:
Partially overlapping substrate spe
- Page 234:
Partially overlapping substrate spe
- Page 238:
Partially overlapping substrate spe
- Page 242:
Partially overlapping substrate spe
- Page 246:
Partially overlapping substrate spe
- Page 250:
Partially overlapping substrate spe
- Page 254:
Partially overlapping substrate spe
- Page 258:
Partially overlapping substrate spe
- Page 262:
Chapter 7 The large mechanosensitiv
- Page 266:
The large mechanosensitive channel
- Page 270:
The large mechanosensitive channel
- Page 274:
The large mechanosensitive channel
- Page 278:
The large mechanosensitive channel
- Page 282:
The large mechanosensitive channel
- Page 286:
The large mechanosensitive channel
- Page 290:
The large mechanosensitive channel
- Page 294:
147
- Page 298:
Chapter 8 The extracellular proteom
- Page 302:
The extracellular proteome of Bacil
- Page 306:
The extracellular proteome of Bacil
- Page 310:
The extracellular proteome of Bacil
- Page 314:
The extracellular proteome of Bacil
- Page 318:
The extracellular proteome of Bacil
- Page 322:
161
- Page 326:
Chapter 9 General summary and discu
- Page 330:
General summary and discussion Pred
- Page 334:
General summary and discussion sugg
- Page 338:
169
- Page 342:
Chapter 10 Reference list 171
- Page 346:
Reference list Berks, B.C., Palmer,
- Page 350:
Reference list Cregg, K.M., Wilding
- Page 354:
Reference list Hartford, O., Franco
- Page 358:
Reference list Kouwen, T.R., Trip,
- Page 362:
Reference list Musser, J.M., Schlie
- Page 366:
Reference list Robinson, C., and Bo
- Page 370:
Reference list Tjalsma, H., Zanen,
- Page 374:
187
- Page 378:
Chapter 11 Nederlandse samenvatting
- Page 382:
Nederlandse samenvatting Staphyloco
- Page 386:
Nederlandse samenvatting genomen di
- Page 390:
Nederlandse samenvatting als lichaa
- Page 394:
Appendix I Dankwoord 197
- Page 398:
Dankwoord onschatbare waarde (Montp
- Page 402:
Dankwoord And last but not least: E
- Page 406:
Appendix II Publication list 203
- Page 410:
Appendices III-V Supplemental table
- Page 414:
Appendix III Supplemental Table III
- Page 418:
Appendix III Supplemental Table III
- Page 422:
Appendix III Supplemental Table III
- Page 426:
Appendix III Supplemental Table III
- Page 430:
Appendix III Supplemental Table III
- Page 434:
Appendix III Supplemental Table III
- Page 438:
Appendix III Supplemental Table III
- Page 442:
Appendix III Supplemental Table III
- Page 446:
Appendix III Supplemental Table III
- Page 450:
Appendix III Supplemental Table III
- Page 454:
Appendix III Supplemental Table III
- Page 458: Appendix III Supplemental Table III
- Page 462: Appendix III Supplemental Table III
- Page 466: Appendix III Supplemental Table III
- Page 470: Appendix IV Supplemental Table IVb
- Page 474: Appendix IV Supplemental Table IVb
- Page 478: Appendix IV Supplemental Table IVb
- Page 482: Appendix IV Supplemental Table IVb
- Page 486: Appendix V Supplemental Table Va Su
- Page 490: Appendix V Supplemental Table Va ID
- Page 494: Appendix V Supplemental Table Va ID
- Page 498: Appendix V Supplemental Table Va ID
- Page 502: Appendix V Supplemental Table Vb Pr
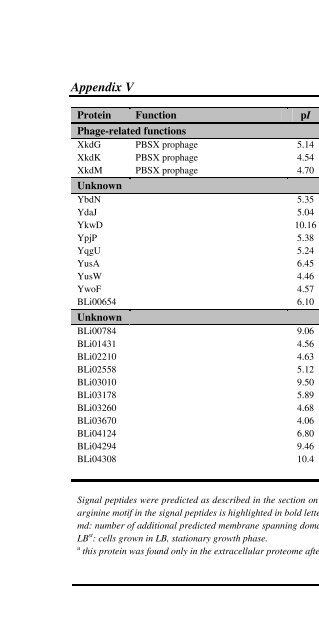
- Page 506: Appendix V Supplemental Table Vb Pr
- Page 512: Appendix V Supplemental Table Vc Pr