Teraflop 73 - Novembre - cesca
Teraflop 73 - Novembre - cesca
Teraflop 73 - Novembre - cesca
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
mildly affected and terminally ill cells,<br />
making it harder to find gene expression<br />
changes. As we currently lack reliable<br />
cellular markers for most neurological<br />
and psychiatric conditions,<br />
there is no obvious way to circumvent<br />
this quandary.<br />
Animal models of disease are an<br />
alternative to the use of human tissue.<br />
These models have had significant<br />
success, particularly in conditions<br />
such as Huntington’s and Alzheimer’s<br />
diseases, for which we have a solid<br />
mechanistic knowledge. But even in<br />
these cases, the genomic analysis has<br />
yet to deliver, as the insights from animal<br />
models are still limited. As some<br />
of the best models involve the insertion<br />
of mutated human genes in the<br />
genome of mice and flies, there might<br />
be unforeseen problems related to<br />
differences in cellular context —all of<br />
the other proteins in the mouse and fly<br />
cells are not human.<br />
In the case of psychiatric disorders,<br />
the situation is even more unfavourable,<br />
as the available models of<br />
these diseases reproduce but their<br />
simplest phenotypes, owing to the fact<br />
that our understanding of the biological<br />
bases of these conditions is quite<br />
elementary. I would therefore argue<br />
that the study of psychiatric diseases<br />
is not yet ripe for genomic analysis.<br />
An embarrassment of riches<br />
Despite the above problems, some<br />
studies have risen to the challenge,<br />
and reported significant and reproducible<br />
gene expression differences<br />
in the brains of several species in<br />
physiological and pathological conditions.<br />
But in addition to showing the<br />
feasibility and value of the genomic<br />
approach, these reports highlight another<br />
of its limitations —the large<br />
number of genes that can be differentially<br />
expressed in a prototypical experiment.<br />
It is common to find that several<br />
dozens of genes are differentially expressed<br />
between two samples. As the<br />
genomic analysis is only the first step<br />
in the characterization of these genes,<br />
we need to confirm these differences<br />
in expression using independent<br />
means before moving forward with<br />
any particular gene. It is clearly im-<br />
practical for any researcher to analyse<br />
in detail every gene, making it necessary<br />
to face the tough decision of<br />
which genes to study. An unfortunate<br />
decision at this stage might lead the<br />
experimenter to a blind alley.<br />
But even assuming that we can<br />
study all the genes that are differentially<br />
expressed, the true problem is how<br />
to make sense of the overwhelming<br />
amount of data that is obtained. If we<br />
find that many synaptic proteins, kinases,<br />
transcription factors and receptors<br />
change between experimental conditions,<br />
will we be able to put all of these<br />
changes into a coherent model of the<br />
process that we are trying to understand?<br />
Is it at all informative that<br />
changes in all those kinds of protein<br />
are found in, say, aging or schizophrenia?<br />
Hardly, I would say. And until we<br />
do not develop a basic understanding<br />
of the core components of the process<br />
that we hope to understand, it will be<br />
very difficult to separate the wheat<br />
from the chaff.<br />
To conclude, I would like to emphasize<br />
that I do not consider that<br />
neurogenomics is a hopeless discipline<br />
that will never yield fruit. But<br />
the problems that I have listed (and<br />
others that I could not discuss for<br />
lack of space) are real, and we must<br />
deal with them so that the brain can<br />
take its rightful place in the postgenomic<br />
era. ■<br />
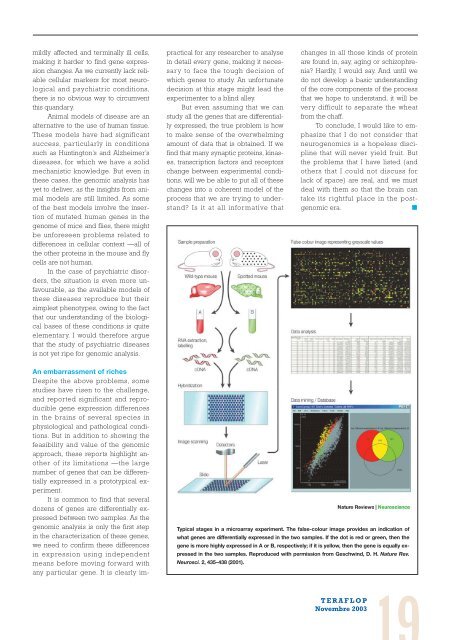
Typical stages in a microarray experiment. The false-colour image provides an indication of<br />
what genes are differentially expressed in the two samples. If the dot is red or green, then the<br />
gene is more highly expressed in A or B, respectively; if it is yellow, then the gene is equally expressed<br />
in the two samples. Reproduced with permission from Geschwind, D. H. Nature Rev.<br />
Neurosci. 2, 435–438 (2001).<br />
TERAFLOP<br />
<strong>Novembre</strong> 2003