FLEISCHWIRTSCHAFT international 1/2017
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
.........................................................................................................<br />
Fleischwirtschaft <strong>international</strong> 1_<strong>2017</strong><br />
51<br />
Research &Development<br />
Sensitivity<br />
PCR-based methods for detecting animal species generallyachieve<br />
detection limits in the range of 0.1% in compound foods (LAUBE,2007;<br />
KÖPPEL,2012). In particular when using universal primer systems, it is<br />
possible that in mixtures of different animal or fish species, the target<br />
fragments are amplified to different extents of efficiency, depending on<br />
the species. This can adverselyaffect the sensitivity of the detection in<br />
certain cases (PIETSCH and WAIBLINGER,2010).<br />
Quantification<br />
Under the conditions described above, multiplex real-time PCR techniques<br />
make it possible to quantify species-specific DNA sequences and<br />
to determine the ratios of these DNA sequences to each other (IWOBI et<br />
al., 2016).<br />
When considering the question of whether acorrelation exists between<br />
DNA ratios and weight ratios of animals in mixed samples, the<br />
following aspects are among those which must be taken into account: in<br />
fatty tissue, compared to meat (muscle tissue), there is generallyless<br />
(around 30%) DNA. Offal such as liver and heart contain 10 to 25 times as<br />
much DNA (originating from mitochondria and the cell nucleus) as muscles<br />
(SCHWÄGELE,2003). When using PCR systems based on target sequences<br />
from mtDNA, it must also be taken into consideration that the<br />
number of mitochondria in animal cells can fluctuate very stronglyfor<br />
example depending on the animal species and tissue (SCHWÄGELE,2003). In<br />
fish roe, mtDNA is the prevailing DNA form (REHBEIN,2003).<br />
ELISA methods<br />
Immunochemical methods such as e.g. Sandwich ELISA (Enzyme Linked<br />
Immunosorbent Assay) or LFD (Lateral Flow Device; Dipsticks) are also<br />
frequentlyused in routine work. Generallykits provided by commercial<br />
suppliers are used. For example, detection methods for the main livestock<br />
species in raw or heated products such as pig/wild boar, cattle/<br />
bison, sheep/goat or equidae (horse, mule, donkey etc.) and deer as a<br />
game species are available. The test kits detect animal species-specific<br />
proteins in both, unheated and heated meat products.<br />
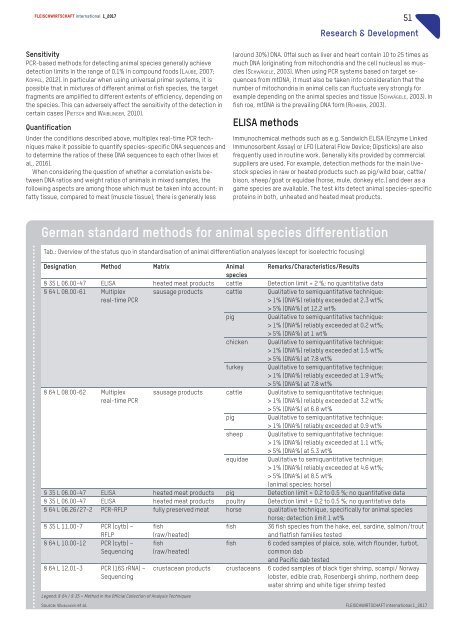
German standard methods for animal species differentiation<br />
Tab.: Overview of the status quo in standardisation of animal differentiation analyses (except for isoelectric focusing)<br />
Designation Method Matrix Animal Remarks/Characteristics/Results<br />
species<br />
§35L06.00-47 ELISA heated meat products cattle Detection limit =2%;noquantitative data<br />
§64L08.00-61 Multiplex<br />
real-time PCR<br />
sausage products cattle Qualitative to semiquantitative technique:<br />
> 1% (DNA%) reliablyexceeded at 2.3 wt%;<br />
> 5% (DNA%) at 12.2 wt%<br />
pig<br />
Qualitative to semiquantitative technique:<br />
> 1% (DNA%) reliablyexceeded at 0.2 wt%;<br />
> 5% (DNA%) at 1wt%<br />
chicken Qualitative to semiquantitative technique:<br />
> 1% (DNA%) reliablyexceeded at 1.5 wt%;<br />
> 5% (DNA%) at 7.8wt%<br />
turkey Qualitative to semiquantitative technique:<br />
> 1% (DNA%) reliablyexceeded at 1.9 wt%;<br />
> 5% (DNA%) at 7.8wt%<br />
§64L08.00-62<br />
Multiplex<br />
real-time PCR<br />
sausage products cattle Qualitative to semiquantitative technique:<br />
> 1% (DNA%) reliablyexceeded at 3.2 wt%;<br />
> 5% (DNA%) at 6.8 wt%<br />
pig<br />
Qualitative to semiquantitative technique:<br />
> 1% (DNA%) reliablyexceeded at 0.9 wt%<br />
sheep Qualitative to semiquantitative technique:<br />
> 1% (DNA%) reliablyexceeded at 1.1 wt%;<br />
> 5% (DNA%) at 5.3 wt%<br />
equidae Qualitative to semiquantitative technique:<br />
> 1% (DNA%) reliablyexceeded at 4.6 wt%;<br />
> 5% (DNA%) at 8.5 wt%<br />
(animal species: horse)<br />
§35L06.00-47 ELISA heated meat products pig Detection limit =0.2 to 0.5 %; no quantitative data<br />
§35L06.00-47 ELISA heated meat products poultry Detection limit =0.2 to 0.5 %; no quantitative data<br />
§64L06.26/27-2 PCR-RFLP fullypreserved meat horse qualitative technique, specificallyfor animal species<br />
horse; detection limit 1wt%<br />
§35L11.00-7 PCR (cytb) –<br />
RFLP<br />
fish<br />
(raw/heated)<br />
fish 36 fish species from the hake, eel, sardine, salmon/trout<br />
and flatfish families tested<br />
§64L10.00-12 PCR (cytb) –<br />
Sequencing<br />
§64L12.01-3 PCR (16S rRNA) –<br />
Sequencing<br />
fish<br />
(raw/heated)<br />
Legend: §64/§35=Methodinthe Official Collection of Analysis Techniques<br />
fish<br />
6coded samples of plaice, sole, witch flounder, turbot,<br />
common dab<br />
and Pacific dab tested<br />
crustacean products crustaceans 6coded samples of black tiger shrimp, scampi/ Norway<br />
lobster, edible crab, Rosenbergii shrimp, northern deep<br />
water shrimp and white tiger shrimp tested<br />
Source: WAIBLINGER et al. <strong>FLEISCHWIRTSCHAFT</strong> <strong>international</strong> 1_<strong>2017</strong>