LIFE09200604007 Tabish - Homi Bhabha National Institute
LIFE09200604007 Tabish - Homi Bhabha National Institute
LIFE09200604007 Tabish - Homi Bhabha National Institute
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Results<br />
4.2.5.2 Real time PCR validation of differentially expressed genes<br />
The genes that were found to be differentially expressed in microarray data<br />
were validated by real time PCR along with GAPDH as an internal control. The genes<br />
included AREG, ARHGAP25, MAFB, MAPK10, TMEFF and ZNF429. Total RNA<br />
isolated from MPN and control cell lines after 5 Gy radiation exposure and 24 h time<br />
point was subjected to cDNA was synthesis which was used for real time validation of<br />
genes. The expression of all the genes was calculated with respect to GAPDH of the<br />
individual samples and a ΔCt value was calculated. This ΔCt value was plotted as<br />
relative fold change by calculating:<br />
Fold change = 2 –ΔCt x 1000<br />
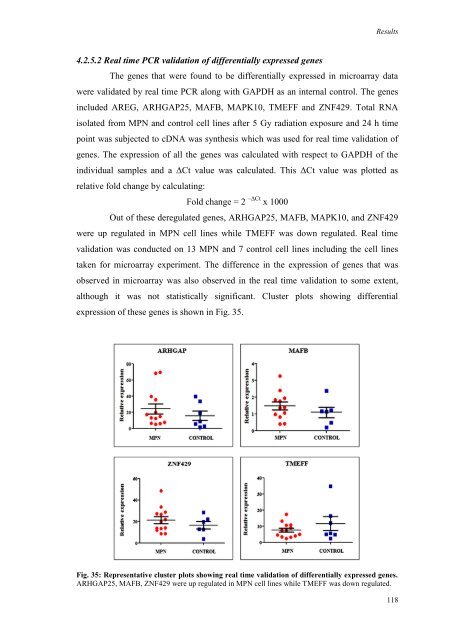
Out of these deregulated genes, ARHGAP25, MAFB, MAPK10, and ZNF429<br />
were up regulated in MPN cell lines while TMEFF was down regulated. Real time<br />
validation was conducted on 13 MPN and 7 control cell lines including the cell lines<br />
taken for microarray experiment. The difference in the expression of genes that was<br />
observed in microarray was also observed in the real time validation to some extent,<br />
although it was not statistically significant. Cluster plots showing differential<br />
expression of these genes is shown in Fig. 35.<br />
Fig. 35: Representative cluster plots showing real time validation of differentially expressed genes.<br />
ARHGAP25, MAFB, ZNF429 were up regulated in MPN cell lines while TMEFF was down regulated.<br />
118