Analysis of microarray data - VSN International
Analysis of microarray data - VSN International
Analysis of microarray data - VSN International
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
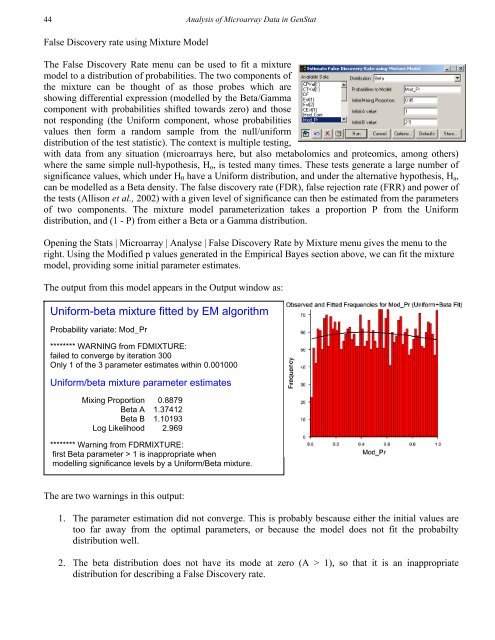
44<strong>Analysis</strong> <strong>of</strong> Microarray Data in GenStatFalse Discovery rate using Mixture ModelThe False Discovery Rate menu can be used to fit a mixturemodel to a distribution <strong>of</strong> probabilities. The two components <strong>of</strong>the mixture can be thought <strong>of</strong> as those probes which areshowing differential expression (modelled by the Beta/Gammacomponent with probabilities shifted towards zero) and thosenot responding (the Uniform component, whose probabilitiesvalues then form a random sample from the null/uniformdistribution <strong>of</strong> the test statistic). The context is multiple testing,with <strong>data</strong> from any situation (<strong>microarray</strong>s here, but also metabolomics and proteomics, among others)where the same simple null-hypothesis, H o , is tested many times. These tests generate a large number <strong>of</strong>significance values, which under H 0 have a Uniform distribution, and under the alternative hypothesis, H a ,can be modelled as a Beta density. The false discovery rate (FDR), false rejection rate (FRR) and power <strong>of</strong>the tests (Allison et al., 2002) with a given level <strong>of</strong> significance can then be estimated from the parameters<strong>of</strong> two components. The mixture model parameterization takes a proportion P from the Uniformdistribution, and (1 - P) from either a Beta or a Gamma distribution.Opening the Stats | Microarray | Analyse | False Discovery Rate by Mixture menu gives the menu to theright. Using the Modified p values generated in the Empirical Bayes section above, we can fit the mixturemodel, providing some initial parameter estimates.The output from this model appears in the Output window as:Uniform-beta mixture fitted by EM algorithmProbability variate: Mod_Pr******** WARNING from FDMIXTURE:failed to converge by iteration 300Only 1 <strong>of</strong> the 3 parameter estimates within 0.001000Uniform/beta mixture parameter estimatesMixing Proportion 0.8879Beta A 1.37412Beta B 1.10193Log Likelihood 2.969******** Warning from FDRMIXTURE:first Beta parameter > 1 is inappropriate whenmodelling significance levels by a Uniform/Beta mixture.The are two warnings in this output:1. The parameter estimation did not converge. This is probably bescause either the initial values aretoo far away from the optimal parameters, or because the model does not fit the probabiltydistribution well.2. The beta distribution does not have its mode at zero (A > 1), so that it is an inappropriatedistribution for describing a False Discovery rate.