nr. 477 - 2011 - Institut for Natur, Systemer og Modeller (NSM)
nr. 477 - 2011 - Institut for Natur, Systemer og Modeller (NSM)
nr. 477 - 2011 - Institut for Natur, Systemer og Modeller (NSM)
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
22 The DuCa Model<br />
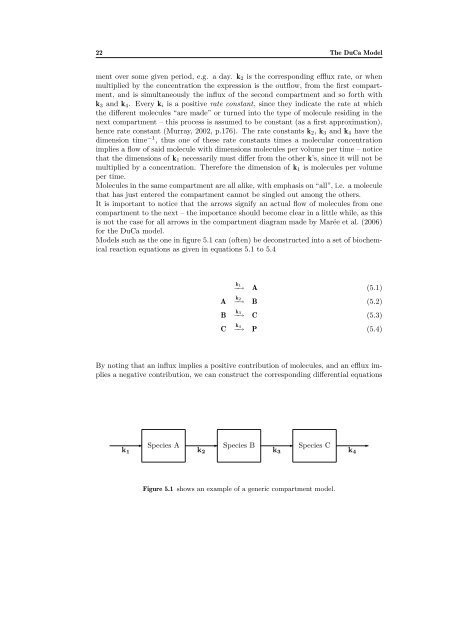
ment over some given period, e.g. a day. k2 is the corresponding efflux rate, or when<br />
multiplied by the concentration the expression is the outflow, from the first compartment,<br />
and is simultaneously the influx of the second compartment and so <strong>for</strong>th with<br />
k3 and k4. Every ki is a positive rate constant, since they indicate the rate at which<br />
the different molecules “are made” or turned into the type of molecule residing in the<br />
next compartment – this process is assumed to be constant (as a first approximation),<br />
hence rate constant (Murray, 2002, p.176). The rate constants k2, k3 and k4 have the<br />
dimension time −1 , thus one of these rate constants times a molecular concentration<br />
implies a flow of said molecule with dimensions molecules per volume per time – notice<br />
that the dimensions of k1 necessarily must differ from the other k’s, since it will not be<br />
multiplied by a concentration. There<strong>for</strong>e the dimension of k1 is molecules per volume<br />
per time.<br />
Molecules in the same compartment are all alike, with emphasis on “all”, i.e. a molecule<br />
that has just entered the compartment cannot be singled out among the others.<br />
It is important to notice that the arrows signify an actual flow of molecules from one<br />
compartment to the next – the importance should become clear in a little while, as this<br />
is not the case <strong>for</strong> all arrows in the compartment diagram made by Marée et al. (2006)<br />
<strong>for</strong> the DuCa model.<br />
Models such as the one in figure 5.1 can (often) be deconstructed into a set of biochemical<br />
reaction equations as given in equations 5.1 to 5.4<br />
k1<br />
−→ A (5.1)<br />
A k2<br />
−→ B (5.2)<br />
B k3<br />
−→ C (5.3)<br />
C k4<br />
−→ P (5.4)<br />
By noting that an influx implies a positive contribution of molecules, and an efflux implies<br />
a negative contribution, we can construct the corresponding differential equations<br />
k1<br />
✲<br />
Species A<br />
k2<br />
✲<br />
Species B<br />
k3<br />
✲<br />
Species C<br />
Figure 5.1 shows an example of a generic compartment model.<br />
k4<br />
✲