Biosafety in the laboratory - VIB
Biosafety in the laboratory - VIB
Biosafety in the laboratory - VIB
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
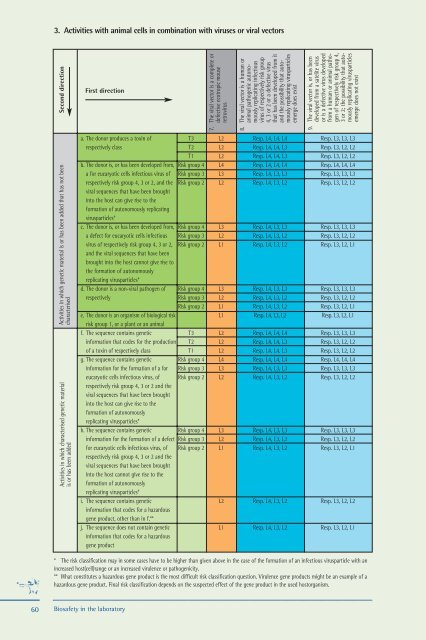
3. Activities with animal cells <strong>in</strong> comb<strong>in</strong>ation with viruses or viral vectors<br />
Second direction<br />
Activities <strong>in</strong> which genetic material is or has been added that has not been<br />
characterised<br />
Activities <strong>in</strong> which characterised genetic material<br />
is or has been added<br />
First direction<br />
7. The viral vector is a complete or<br />
defective ecotropic mouse<br />
retrovirus<br />
8. The viral vector is a human or<br />
animal pathogenic autonomously<br />
replicat<strong>in</strong>g <strong>in</strong>fectious<br />
virus of respectively risk group<br />
4, 3 or 2 or a defective virus<br />
that has been developed from it<br />
and <strong>the</strong> possibility that automously<br />
replicat<strong>in</strong>g virusparticles<br />
emerge does exist<br />
9. The viral vector is, or has been<br />
developed from a satelite virus<br />
or is a defective virus developed<br />
from a human or animal pathogen<br />
of respectively risk group 4,<br />
3 or 2; <strong>the</strong> possibility that automously<br />
replicat<strong>in</strong>g virusparticles<br />
emerge does not exist<br />
a. The donor produces a tox<strong>in</strong> of T3 L2 Resp. L4, L4, L4 Resp. L3, L3, L3<br />
respectively class T2 L2 Resp. L4, L4, L3 Resp. L3, L2, L2<br />
T1 L2 Resp. L4, L4, L3 Resp. L3, L2, L2<br />
b. The donor is, or has been developed from, Risk group 4 L4 Resp. L4, L4, L4 Resp. L4, L4, L4<br />
a for eucaryotic cells <strong>in</strong>fectious virus of Risk group 3 L3 Resp. L4, L3, L3 Resp. L3, L3, L3<br />
respectively risk group 4, 3 or 2, and <strong>the</strong> Risk group 2 L2 Resp. L4, L3, L2 Resp. L3, L2, L2<br />
viral sequences that have been brought<br />
<strong>in</strong>to <strong>the</strong> host can give rise to <strong>the</strong><br />
formation of autonomously replicat<strong>in</strong>g<br />
virusparticles*<br />
c. The donor is, or has been developed from, Risk group 4 L3 Resp. L4, L3, L3 Resp. L3, L3, L3<br />
a defect for eucaryotic cells <strong>in</strong>fectious Risk group 3 L2 Resp. L4, L3, L2 Resp. L3, L2, L2<br />
virus of respectively risk group 4, 3 or 2, Risk group 2 L1 Resp. L4, L3, L2 Resp. L3, L2, L1<br />
and <strong>the</strong> viral sequences that have been<br />
brought <strong>in</strong>to <strong>the</strong> host cannot give rise to<br />
<strong>the</strong> formation of autonomously<br />
replicat<strong>in</strong>g virusparticles*<br />
d. The donor is a non-viral pathogen of Risk group 4 L3 Resp. L4, L3, L3 Resp. L3, L3, L3<br />
respectively Risk group 3 L2 Resp. L4, L3, L2 Resp. L3, L2, L2<br />
Risk group 2 L1 Resp. L4, L3, L2 Resp. L3, L2, L1<br />
e. The donor is an organism of biological risk L1 Resp. L4, L3, L2 Resp. L3, L2, L1<br />
risk group 1, or a plant or an animal<br />
f. The sequence conta<strong>in</strong>s genetic T3 L2 Resp. L4, L4, L4 Resp. L3, L3, L3<br />
<strong>in</strong>formation that codes for <strong>the</strong> production T2 L2 Resp. L4, L4, L3 Resp. L3, L2, L2<br />
of a tox<strong>in</strong> of respectively class T1 L2 Resp. L4, L4, L3 Resp. L3, L2, L2<br />
g. The sequence conta<strong>in</strong>s genetic Risk group 4 L4 Resp. L4, L4, L4 Resp. L4, L4, L4<br />
<strong>in</strong>formation for <strong>the</strong> formation of a for Risk group 3 L3 Resp. L4, L3, L3 Resp. L3, L3, L3<br />
eucaryotic cells <strong>in</strong>fectious virus, of Risk group 2 L2 Resp. L4, L3, L2 Resp. L3, L2, L2<br />
respectively risk group 4, 3 or 2 and <strong>the</strong><br />
viral sequences that have been brought<br />
<strong>in</strong>to <strong>the</strong> host can give rise to <strong>the</strong><br />
formation of autonomously<br />
replicat<strong>in</strong>g virusparticles*<br />
h. The sequence conta<strong>in</strong>s genetic Risk group 4 L3 Resp. L4, L3, L3 Resp. L3, L3, L3<br />
<strong>in</strong>formation for <strong>the</strong> formation of a defect Risk group 3 L2 Resp. L4, L3, L2 Resp. L3, L2, L2<br />
for eucaryotic cells <strong>in</strong>fectious virus, of Risk group 2 L1 Resp. L4, L3, L2 Resp. L3, L2, L1<br />
respectively risk group 4, 3 or 2 and <strong>the</strong><br />
viral sequences that have been brought<br />
<strong>in</strong>to <strong>the</strong> host cannot give rise to <strong>the</strong><br />
formation of autonomously<br />
replicat<strong>in</strong>g virusparticles*<br />
i. The sequence conta<strong>in</strong>s genetic L2 Resp. L4, L3, L2 Resp. L3, L2, L2<br />
<strong>in</strong>formation that codes for a hazardous<br />
gene product, o<strong>the</strong>r than <strong>in</strong> f.**<br />
j. The sequence does not conta<strong>in</strong> genetic L1 Resp. L4, L3, L2 Resp. L3, L2, L1<br />
<strong>in</strong>formation that codes for a hazardous<br />
gene product<br />
* The risk classification may <strong>in</strong> some cases have to be higher than given above <strong>in</strong> <strong>the</strong> case of <strong>the</strong> formation of an <strong>in</strong>fectious virusparticle with an<br />
<strong>in</strong>creased host(cell)range or an <strong>in</strong>creased virulence or pathogenicity.<br />
** What constitutes a hazardous gene product is <strong>the</strong> most difficult risk classification question. Virulence gene products might be an example of a<br />
hazardous gene product. F<strong>in</strong>al risk classification depends on <strong>the</strong> suspected effect of <strong>the</strong> gene product <strong>in</strong> <strong>the</strong> used hostorganism.<br />
60<br />
<strong>Biosafety</strong> <strong>in</strong> <strong>the</strong> <strong>laboratory</strong>