Theoretical and Experimental DNA Computation (Natural ...
Theoretical and Experimental DNA Computation (Natural ...
Theoretical and Experimental DNA Computation (Natural ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
120 5 Physical Implementations<br />
3. To test the error-resistance of a sequence of removal operations that would<br />
occur during an actual algorithmic implementation.<br />
Encoding colorings in <strong>DNA</strong><br />
We construct an initial library of (not necessarily legal) colorings in the following<br />
manner. For each vertex vi ∈ V we synthesize a single oligonucleotide (or<br />
oligo) to represent each of vi =red,vi = green, <strong>and</strong> vi = blue. The structure<br />
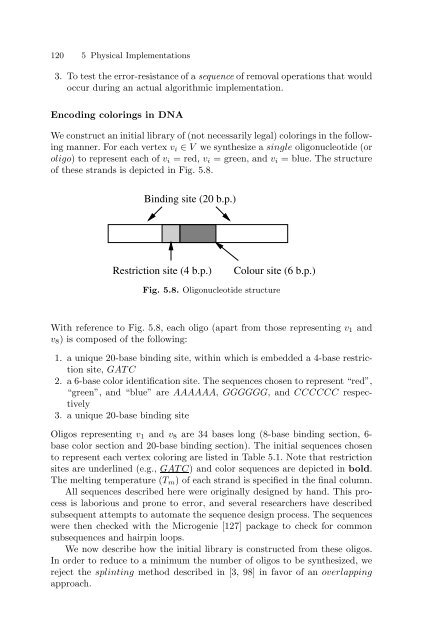
of these str<strong>and</strong>s is depicted in Fig. 5.8.<br />
Binding site (20 b.p.)<br />
Restriction site (4 b.p.) Colour site (6 b.p.)<br />
Fig. 5.8. Oligonucleotide structure<br />
With reference to Fig. 5.8, each oligo (apart from those representing v1 <strong>and</strong><br />
v8) is composed of the following:<br />
1. a unique 20-base binding site, within which is embedded a 4-base restriction<br />
site, GAT C<br />
2. a 6-base color identification site. The sequences chosen to represent “red”,<br />
“green”, <strong>and</strong> “blue” are AAAAAA, GGGGGG, <strong>and</strong>CCCCCC respectively<br />
3. a unique 20-base binding site<br />
Oligos representing v1 <strong>and</strong> v8 are 34 bases long (8-base binding section, 6base<br />
color section <strong>and</strong> 20-base binding section). The initial sequences chosen<br />
to represent each vertex coloring are listed in Table 5.1. Note that restriction<br />
sites are underlined (e.g., GAT C) <strong>and</strong> color sequences are depicted in bold.<br />
The melting temperature (Tm) of each str<strong>and</strong> is specified in the final column.<br />
All sequences described here were originally designed by h<strong>and</strong>. This process<br />
is laborious <strong>and</strong> prone to error, <strong>and</strong> several researchers have described<br />
subsequent attempts to automate the sequence design process. The sequences<br />
were then checked with the Microgenie [127] package to check for common<br />
subsequences <strong>and</strong> hairpin loops.<br />
We now describe how the initial library is constructed from these oligos.<br />
In order to reduce to a minimum the number of oligos to be synthesized, we<br />
reject the splinting method described in [3, 98] in favor of an overlapping<br />
approach.