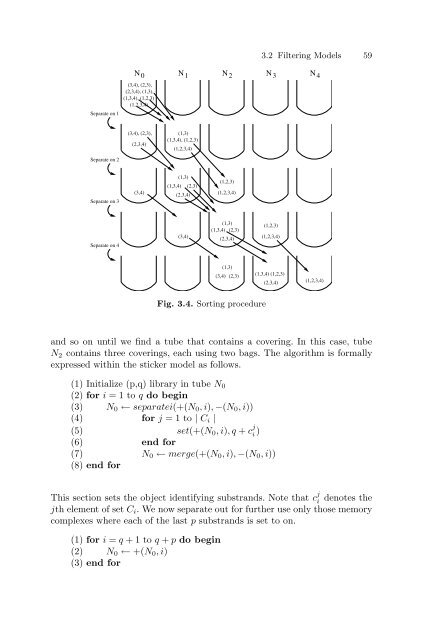
Separate on 1 Separate on 2 Separate on 3 Separate on 4 (3,4), (2,3), (2,3,4), (1,3), (1,3,4), (1,2,3) (1,2,3,4) (3,4), (2,3), (2,3,4) (3,4) (1,3) (1,3,4), (1,2,3) (1,2,3,4) (1,3,4) (1,3) (2,3,4) (3,4) (2,3) (1,2,3) (1,2,3,4) (1,3) (1,3,4) (2,3) (2,3,4) (1,3) 3.2 Filtering Models 59 N0 N1 N2 N3 N4 (3,4) (2,3) Fig. 3.4. Sorting procedure (1,2,3) (1,2,3,4) (1,3,4) (1,2,3) (2,3,4) (1,2,3,4) <strong>and</strong> so on until we find a tube that contains a covering. In this case, tube N2 contains three coverings, each using two bags. The algorithm is formally expressed within the sticker model as follows. (1) Initialize (p,q) library in tube N0 (2) for i =1toq do begin (3) N0 ← separatei(+(N0,i), −(N0,i)) (4) for j =1to| Ci | (5) set(+(N0,i),q+ c j i ) (6) end for (7) N0 ← merge(+(N0,i), −(N0,i)) (8) end for This section sets the object identifying substr<strong>and</strong>s. Note that c j i denotes the jth element of set Ci. We now separate out for further use only those memory complexes where each of the last p substr<strong>and</strong>s is set to on. (1) for i = q +1toq + p do begin (2) N0 ← +(N0,i) (3) end for
60 3 Models of Molecular <strong>Computation</strong> We now sort the remaining str<strong>and</strong>s according to how many bags they encode. (1) for i =0toq − 1 do begin (2) for j − 1 down to 0 do begin (3) separate(+(Nj,i+1), −(Nj,i+1)) (4) Nj+1 ← merge(+(Nj,i+1),Nj+1) (5) Nj ←−(Nj,i+1) (6) end for (7) end for Line 3 separates each tube according to the value of i, <strong>and</strong> line 4 performs the right shift of selected str<strong>and</strong>s. We then search for a final output: (1) Read N1 (2) else if empty then read N2 (3) else if empty then read N3 (4) ... 3.3 Splicing Models Since any instance of any problem in the complexity class NP may be expressed in terms of an instance of any NP-complete problem, it follows that the multi-set operations described earlier at least implicitly provide sufficient computational power to solve any problem in NP. We do not believe they provide the full algorithmic computational power of a Turing Machine. Without the availability of string editing operations, it is difficult to see how the transition from one state of the Turing Machine to another may be achieved using <strong>DNA</strong>. However, as several authors have recently described, one further operation, the so-called splicing operation, will provide full Turing computability. Here we provide an overview of the various splicing models proposed. Let S <strong>and</strong> T be two strings over the alphabet α. Then the splice operation consists of cutting S <strong>and</strong> T at specific positions <strong>and</strong> concatenating the resulting prefix of S with the suffix of T <strong>and</strong> concatenating the prefix of T with the suffix of S (Fig. 3.5). This operation is similar to the crossover operation employed by genetic algorithms [68, 96]. Splicing systems date back to 1987, with the publication of Tom Head’s seminal paper [77] (see also [78]). In [50], the authors show that the generative power of finite extended splicing systems is equal to that of Turing Machines. For an excellent review of splicing systems, the reader is directed to [120]. Reif’s PAM model In [128], Reif within his so-called Parallel Associative Memory Model describes a Parallel Associative Matching (PA-Match) operation. The essential con-
- Page 2 and 3:
Natural Computing Series Series Edi
- Page 4:
Martyn Amos Department of Computer
- Page 8 and 9:
Preface DNA computation has emerged
- Page 10:
Preface IX acknowledged. Finally, I
- Page 13 and 14:
XII Contents 4 Complexity Issues ..
- Page 16 and 17:
Introduction “Where a calculator
- Page 18 and 19:
Introduction 3 Even though their un
- Page 20 and 21:
1 DNA: The Molecule of Life “All
- Page 22 and 23:
1.3 DNA as the Carrier of Genetic I
- Page 24 and 25: 1.3 DNA as the Carrier of Genetic I
- Page 26 and 27: Synthesis 1.4 Operations on DNA 11
- Page 28 and 29: Gel electrophoresis 1.4 Operations
- Page 30 and 31: 5’ 3’ A T A G A G T T 3’ TCA
- Page 32 and 33: 1.4 Operations on DNA 17 Others may
- Page 34 and 35: Vector Strand to be inserted (targe
- Page 36: 1.5 Summary 1.6 Bibliographical Not
- Page 39 and 40: 24 2 Theoretical Computer Science:
- Page 41 and 42: 26 2 Theoretical Computer Science:
- Page 43 and 44: 28 2 Theoretical Computer Science:
- Page 45 and 46: 30 2 Theoretical Computer Science:
- Page 47 and 48: 32 2 Theoretical Computer Science:
- Page 49 and 50: 34 2 Theoretical Computer Science:
- Page 51 and 52: 36 2 Theoretical Computer Science:
- Page 53 and 54: 38 2 Theoretical Computer Science:
- Page 55 and 56: 40 2 Theoretical Computer Science:
- Page 57 and 58: 42 2 Theoretical Computer Science:
- Page 59 and 60: 44 2 Theoretical Computer Science:
- Page 61 and 62: 46 3 Models of Molecular Computatio
- Page 63 and 64: 48 3 Models of Molecular Computatio
- Page 65 and 66: 50 3 Models of Molecular Computatio
- Page 67 and 68: 52 3 Models of Molecular Computatio
- Page 69 and 70: 54 3 Models of Molecular Computatio
- Page 71 and 72: 56 3 Models of Molecular Computatio
- Page 73: 58 3 Models of Molecular Computatio
- Page 77 and 78: 62 3 Models of Molecular Computatio
- Page 79 and 80: 64 3 Models of Molecular Computatio
- Page 81 and 82: 66 3 Models of Molecular Computatio
- Page 83 and 84: 68 3 Models of Molecular Computatio
- Page 85 and 86: 70 3 Models of Molecular Computatio
- Page 87 and 88: 72 4 Complexity Issues is that, alt
- Page 89 and 90: 74 4 Complexity Issues The weak par
- Page 91 and 92: 76 4 Complexity Issues 4.3 A Strong
- Page 93 and 94: 78 4 Complexity Issues Ω = {∧,
- Page 95 and 96: 80 4 Complexity Issues 0 1 0 1 x1 x
- Page 97 and 98: 82 4 Complexity Issues connecting a
- Page 99 and 100: 84 4 Complexity Issues Level simula
- Page 101 and 102: 86 4 Complexity Issues At level D(S
- Page 103 and 104: 88 4 Complexity Issues xANDy)) and
- Page 105 and 106: 90 4 Complexity Issues Input matrix
- Page 107 and 108: 92 4 Complexity Issues memory acces
- Page 109 and 110: 94 4 Complexity Issues denoted by P
- Page 111 and 112: 96 4 Complexity Issues The final st
- Page 113 and 114: 98 4 Complexity Issues 1)), ADDR[])
- Page 115 and 116: 100 4 Complexity Issues Size(STI)=O
- Page 117 and 118: 102 4 Complexity Issues best attain
- Page 119 and 120: 104 4 Complexity Issues 10. LDC 0 1
- Page 121 and 122: 106 4 Complexity Issues ACC[] := bi
- Page 124 and 125:
5 Physical Implementations “No am
- Page 126 and 127:
5.3 Initial Set Construction Within
- Page 128 and 129:
Vertex 1 Vertex 2 Vertex 3 ¡ ¡ ¡
- Page 130 and 131:
5.5 Evaluation of Adleman’s Imple
- Page 132 and 133:
5.6 Implementation of the Parallel
- Page 134 and 135:
5.8 Experimental Investigations 119
- Page 136 and 137:
5.8 Experimental Investigations 121
- Page 138 and 139:
Create initial strand library Confi
- Page 140 and 141:
5.8 Experimental Investigations 125
- Page 142 and 143:
5.8 Experimental Investigations 127
- Page 144 and 145:
Experiment 25. New full-length chai
- Page 146 and 147:
5.8 Experimental Investigations 131
- Page 148 and 149:
5.8 Experimental Investigations 133
- Page 150 and 151:
5.9 Other Laboratory Implementation
- Page 152 and 153:
5.9 Other Laboratory Implementation
- Page 154 and 155:
5.9 Other Laboratory Implementation
- Page 156 and 157:
5.9 Other Laboratory Implementation
- Page 158 and 159:
5.9 Other Laboratory Implementation
- Page 160:
5.11 Bibliographical Notes 145 whic
- Page 163 and 164:
148 6 Cellular Computing of truly i
- Page 165 and 166:
150 6 Cellular Computing 6.2 Succes
- Page 167 and 168:
152 6 Cellular Computing are “glu
- Page 169 and 170:
154 6 Cellular Computing x y z x (a
- Page 171 and 172:
156 6 Cellular Computing 6.7 Biblio
- Page 173 and 174:
158 References 14. Martyn Amos, Ste
- Page 175 and 176:
160 References 53. Dónall A. Mac D
- Page 177 and 178:
162 References 92. D. Knuth. The Ar
- Page 179 and 180:
164 References 135. Kensaku Sakamot
- Page 182 and 183:
Index ALGOL, 102 AND, 23-24, 42, 87
- Page 184 and 185:
iological, 74, 114, 150 Feynman, R.
- Page 186 and 187:
Petrinet,63 phage, 18-20 phosphate,
- Page 188:
Natural Computing Series W.M. Spear