New Statistical Algorithms for the Analysis of Mass - FU Berlin, FB MI ...
New Statistical Algorithms for the Analysis of Mass - FU Berlin, FB MI ...
New Statistical Algorithms for the Analysis of Mass - FU Berlin, FB MI ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
1.2. GOALS, OBJECTIVES AND TASKS 7<br />
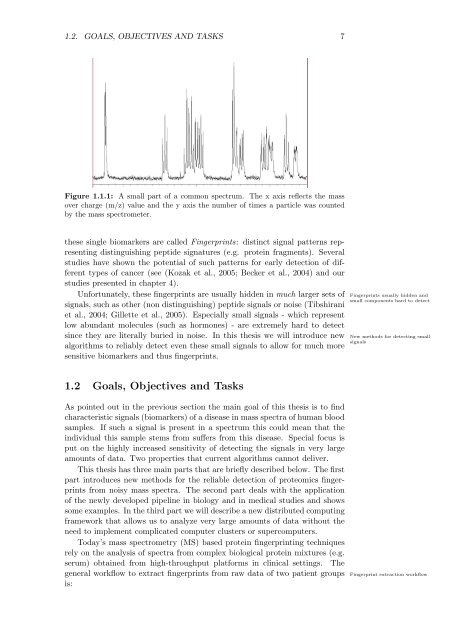
Figure 1.1.1: A small part <strong>of</strong> a common spectrum. The x axis reflects <strong>the</strong> mass<br />
over charge (m/z) value and <strong>the</strong> y axis <strong>the</strong> number <strong>of</strong> times a particle was counted<br />
by <strong>the</strong> mass spectrometer.<br />
<strong>the</strong>se single biomarkers are called Fingerprints: distinct signal patterns representing<br />
distinguishing peptide signatures (e.g. protein fragments). Several<br />
studies have shown <strong>the</strong> potential <strong>of</strong> such patterns <strong>for</strong> early detection <strong>of</strong> different<br />
types <strong>of</strong> cancer (see (Kozak et al., 2005; Becker et al., 2004) and our<br />
studies presented in chapter 4).<br />
Un<strong>for</strong>tunately, <strong>the</strong>se fingerprints are usually hidden in much larger sets <strong>of</strong> Fingerprints usually hidden and<br />
small components hard to detect<br />
signals, such as o<strong>the</strong>r (non distinguishing) peptide signals or noise (Tibshirani<br />
et al., 2004; Gillette et al., 2005). Especially small signals - which represent<br />
low abundant molecules (such as hormones) - are extremely hard to detect<br />
since <strong>the</strong>y are literally buried in noise. In this <strong>the</strong>sis we will introduce new <strong>New</strong> methods <strong>for</strong> detecting small<br />
signals<br />
algorithms to reliably detect even <strong>the</strong>se small signals to allow <strong>for</strong> much more<br />
sensitive biomarkers and thus fingerprints.<br />
1.2 Goals, Objectives and Tasks<br />
As pointed out in <strong>the</strong> previous section <strong>the</strong> main goal <strong>of</strong> this <strong>the</strong>sis is to find<br />
characteristic signals (biomarkers) <strong>of</strong> a disease in mass spectra <strong>of</strong> human blood<br />
samples. If such a signal is present in a spectrum this could mean that <strong>the</strong><br />
individual this sample stems from suffers from this disease. Special focus is<br />
put on <strong>the</strong> highly increased sensitivity <strong>of</strong> detecting <strong>the</strong> signals in very large<br />
amounts <strong>of</strong> data. Two properties that current algorithms cannot deliver.<br />
This <strong>the</strong>sis has three main parts that are briefly described below. The first<br />
part introduces new methods <strong>for</strong> <strong>the</strong> reliable detection <strong>of</strong> proteomics fingerprints<br />
from noisy mass spectra. The second part deals with <strong>the</strong> application<br />
<strong>of</strong> <strong>the</strong> newly developed pipeline in biology and in medical studies and shows<br />
some examples. In <strong>the</strong> third part we will describe a new distributed computing<br />
framework that allows us to analyze very large amounts <strong>of</strong> data without <strong>the</strong><br />
need to implement complicated computer clusters or supercomputers.<br />
Today’s mass spectrometry (MS) based protein fingerprinting techniques<br />
rely on <strong>the</strong> analysis <strong>of</strong> spectra from complex biological protein mixtures (e.g.<br />
serum) obtained from high-throughput plat<strong>for</strong>ms in clinical settings. The<br />
general workflow to extract fingerprints from raw data <strong>of</strong> two patient groups Fingerprint extraction workflow<br />
is: