Research Report 2010 2011 - Helmholtz-Zentrum für ...
Research Report 2010 2011 - Helmholtz-Zentrum für ...
Research Report 2010 2011 - Helmholtz-Zentrum für ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
58 SPECIAL FEATURES | Nuclear Magnetic Resonance Spectroscopy, a Major Player in the Arsenal of Platform Technologies<br />
Large proteins and the compatibility with X-ray<br />
The question now arises as to whether NMR spectroscopy<br />
can also provide structural details for larger proteins and<br />
are the structures determined in solution compatible with<br />
those in the solid phase from X-ray crystallography. In<br />
general, the answer to both questions is yes. Laboratories<br />
such as the HZI Protein Sample Production Facility can now<br />
produce 15 N, 13 C-doubly-labelled protein using recombinant<br />
methods suitable for multi-dimensional heteronuclear NMR<br />
investigations in solution. A salient example is the elucidation<br />
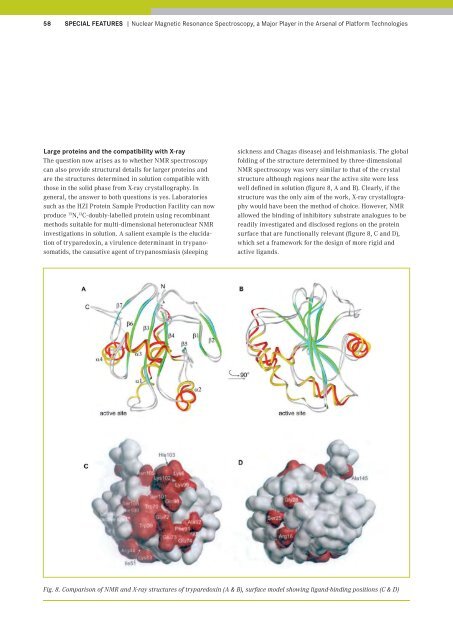
of tryparedoxin, a virulence determinant in trypanosomatids,<br />
the causative agent of trypanosmiasis (sleeping<br />
sickness and Chagas disease) and leishmaniasis. The global<br />
folding of the structure determined by three-dimensional<br />
NMR spectroscopy was very similar to that of the crystal<br />
structure although regions near the active site were less<br />
well defined in solution (figure 8, A and B). Clearly, if the<br />
structure was the only aim of the work, X-ray crystallography<br />
would have been the method of choice. However, NMR<br />
allowed the binding of inhibitory substrate analogues to be<br />
readily investigated and disclosed regions on the protein<br />
surface that are functionally relevant (figure 8, C and D),<br />
which set a framework for the design of more rigid and<br />
active ligands.<br />
Fig. 8. Comparison of NMR and X-ray structures of tryparedoxin (A & B), surface model showing ligand-binding positions (C & D)