5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
3.1 Capture experiments <strong>in</strong>volv<strong>in</strong>g genome sequenced bacterial<br />
stra<strong>in</strong>s<br />
This section will describe <strong>the</strong> <strong>in</strong>itial capture experiments us<strong>in</strong>g reference genomes.<br />
<strong>The</strong> experiments were designed to develop <strong>the</strong> methodology associated with<br />
genomic island capture technique and to test <strong>the</strong> size limits <strong>of</strong> <strong>the</strong> captured <strong>in</strong>sert.<br />
For E. coli, <strong>the</strong> reference genome was K12 MG1655 and for P. aerug<strong>in</strong>osa PAO1<br />
and PA14.<br />
<strong>The</strong> yeast cell spheroplasts are transformed with <strong>the</strong> capture vector plus <strong>the</strong> genomic<br />
DNA. Homologous recomb<strong>in</strong>ation between <strong>the</strong> capture vector and <strong>the</strong> genomic<br />
DNA results <strong>in</strong> <strong>the</strong> genomic island now be<strong>in</strong>g present with<strong>in</strong> <strong>the</strong> vector. <strong>The</strong> ma<strong>in</strong><br />
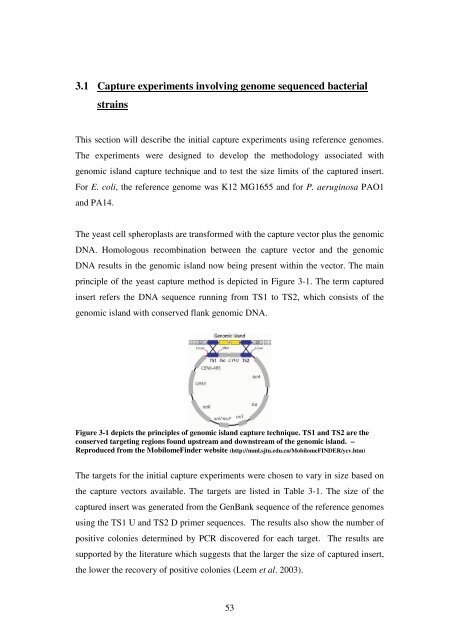
pr<strong>in</strong>ciple <strong>of</strong> <strong>the</strong> yeast capture method is depicted <strong>in</strong> Figure 3-1. <strong>The</strong> term captured<br />
<strong>in</strong>sert refers <strong>the</strong> DNA sequence runn<strong>in</strong>g from TS1 to TS2, which consists <strong>of</strong> <strong>the</strong><br />
genomic island with conserved flank genomic DNA.<br />
Figure 3-1 depicts <strong>the</strong> pr<strong>in</strong>ciples <strong>of</strong> genomic island capture technique. TS1 and TS2 are <strong>the</strong><br />
conserved target<strong>in</strong>g regions found upstream and downstream <strong>of</strong> <strong>the</strong> genomic island. –<br />
Reproduced from <strong>the</strong> MobilomeF<strong>in</strong>der website (http://mml.sjtu.edu.cn/MobilomeFINDER/ycv.htm)<br />
<strong>The</strong> targets for <strong>the</strong> <strong>in</strong>itial capture experiments were chosen to vary <strong>in</strong> size based on<br />
<strong>the</strong> capture vectors available. <strong>The</strong> targets are listed <strong>in</strong> Table 3-1. <strong>The</strong> size <strong>of</strong> <strong>the</strong><br />
captured <strong>in</strong>sert was generated from <strong>the</strong> GenBank sequence <strong>of</strong> <strong>the</strong> reference genomes<br />
us<strong>in</strong>g <strong>the</strong> TS1 U and TS2 D primer sequences. <strong>The</strong> results also show <strong>the</strong> number <strong>of</strong><br />
positive colonies determ<strong>in</strong>ed by PCR discovered for each target. <strong>The</strong> results are<br />
supported by <strong>the</strong> literature which suggests that <strong>the</strong> larger <strong>the</strong> size <strong>of</strong> captured <strong>in</strong>sert,<br />
<strong>the</strong> lower <strong>the</strong> recovery <strong>of</strong> positive colonies (Leem et al. 2003).<br />
53