5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
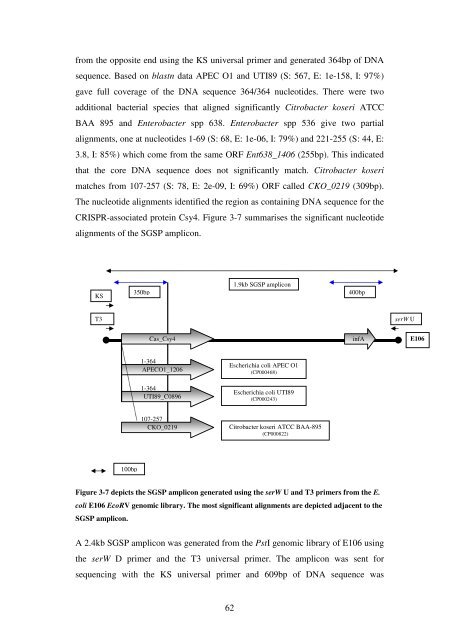
from <strong>the</strong> opposite end us<strong>in</strong>g <strong>the</strong> KS universal primer and generated 364bp <strong>of</strong> DNA<br />
sequence. Based on blastn data APEC O1 and UTI89 (S: 567, E: 1e-158, I: 97%)<br />
gave full coverage <strong>of</strong> <strong>the</strong> DNA sequence 364/364 nucleotides. <strong>The</strong>re were two<br />
additional bacterial species that aligned significantly Citrobacter koseri ATCC<br />
BAA 895 and Enterobacter spp 638. Enterobacter spp 536 give two partial<br />
alignments, one at nucleotides 1-69 (S: 68, E: 1e-06, I: 79%) and 221-255 (S: 44, E:<br />
3.8, I: 85%) which come from <strong>the</strong> same ORF Ent638_1406 (255bp). This <strong>in</strong>dicated<br />
that <strong>the</strong> core DNA sequence does not significantly match. Citrobacter koseri<br />
matches from 107-257 (S: 78, E: 2e-09, I: 69%) ORF called CKO_0219 (309bp).<br />
<strong>The</strong> nucleotide alignments identified <strong>the</strong> region as conta<strong>in</strong><strong>in</strong>g DNA sequence for <strong>the</strong><br />
CRISPR-associated prote<strong>in</strong> Csy4. Figure 3-7 summarises <strong>the</strong> significant nucleotide<br />
alignments <strong>of</strong> <strong>the</strong> SGSP amplicon.<br />
KS<br />
T3<br />
Figure 3-7 depicts <strong>the</strong> SGSP amplicon generated us<strong>in</strong>g <strong>the</strong> serW U and T3 primers from <strong>the</strong> E.<br />
coli E106 EcoRV genomic library. <strong>The</strong> most significant alignments are depicted adjacent to <strong>the</strong><br />
SGSP amplicon.<br />
100bp<br />
1.9kb SGSP amplicon<br />
350bp 400bp<br />
Cas_Csy4 <strong>in</strong>fA<br />
1-364<br />
APECO1_1206<br />
1-364<br />
UTI89_C0896<br />
Escherichia coli APEC O1<br />
(CP000468)<br />
107-257<br />
CKO_0219 Citrobacter koseri ATCC BAA-895<br />
(CP000822)<br />
A 2.4kb SGSP amplicon was generated from <strong>the</strong> PstI genomic library <strong>of</strong> E106 us<strong>in</strong>g<br />
<strong>the</strong> serW D primer and <strong>the</strong> T3 universal primer. <strong>The</strong> amplicon was sent for<br />
sequenc<strong>in</strong>g with <strong>the</strong> KS universal primer and 609bp <strong>of</strong> DNA sequence was<br />
62<br />
Escherichia coli UTI89<br />
(CP000243)<br />
serW U<br />
E106