5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
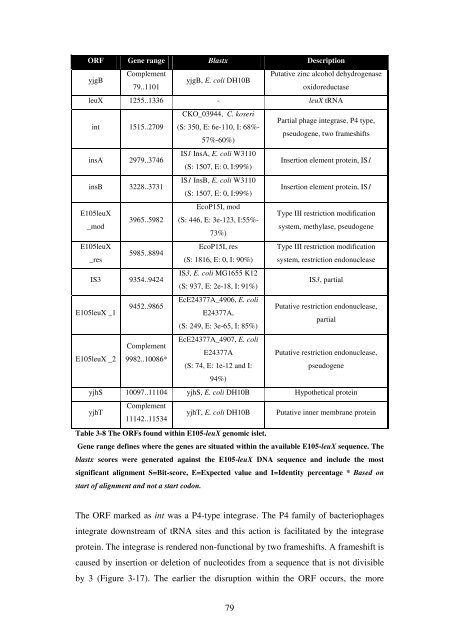
ORF Gene range Blastx Description<br />
yjgB<br />
Complement<br />
79..1101<br />
yjgB, E. coli DH10B<br />
79<br />
Putative z<strong>in</strong>c alcohol dehydrogenase<br />
oxidoreductase<br />
leuX 1255..1336 - leuX tRNA<br />
<strong>in</strong>t 1515..2709<br />
<strong>in</strong>sA 2979..3746<br />
<strong>in</strong>sB 3228..3731<br />
E105leuX<br />
_mod<br />
E105leuX<br />
_res<br />
3965..5982<br />
5985..8894<br />
IS3 9354..9424<br />
E105leuX _1<br />
E105leuX _2<br />
9452..9865<br />
Complement<br />
9982..10086*<br />
CKO_03944, C. koseri<br />
(S: 350, E: 6e-110, I: 68%-<br />
57%-60%)<br />
IS1 InsA, E. coli W3110<br />
(S: 1507, E: 0, I:99%)<br />
IS1 InsB, E. coli W3110<br />
(S: 1507, E: 0, I:99%)<br />
EcoP15I, mod<br />
(S: 446, E: 3e-123, I:55%-<br />
73%)<br />
EcoP15I, res<br />
(S: 1816, E: 0, I: 90%)<br />
IS3, E. coli MG1655 K12<br />
(S: 937, E: 2e-18, I: 91%)<br />
EcE24377A_4906, E. coli<br />
E24377A,<br />
(S: 249, E: 3e-65, I: 85%)<br />
EcE24377A_4907, E. coli<br />
E24377A<br />
(S: 74, E: 1e-12 and I:<br />
94%)<br />
Partial phage <strong>in</strong>tegrase, P4 type,<br />
pseudogene, two frameshifts<br />
Insertion element prote<strong>in</strong>, IS1<br />
Insertion element prote<strong>in</strong>, IS1<br />
Type III restriction modification<br />
system, methylase, pseudogene<br />
Type III restriction modification<br />
system, restriction endonuclease<br />
IS3, partial<br />
Putative restriction endonuclease,<br />
partial<br />
Putative restriction endonuclease,<br />
pseudogene<br />
yjhS 10097..11104 yjhS, E. coli DH10B Hypo<strong>the</strong>tical prote<strong>in</strong><br />
yjhT<br />
Complement<br />
11142..11534<br />
Table 3-8 <strong>The</strong> ORFs found with<strong>in</strong> E105-leuX genomic islet.<br />
yjhT, E. coli DH10B Putative <strong>in</strong>ner membrane prote<strong>in</strong><br />
Gene range def<strong>in</strong>es where <strong>the</strong> genes are situated with<strong>in</strong> <strong>the</strong> available E105-leuX sequence. <strong>The</strong><br />
blastx scores were generated aga<strong>in</strong>st <strong>the</strong> E105-leuX DNA sequence and <strong>in</strong>clude <strong>the</strong> most<br />
significant alignment S=Bit-score, E=Expected value and I=Identity percentage * Based on<br />
start <strong>of</strong> alignment and not a start codon.<br />
<strong>The</strong> ORF marked as <strong>in</strong>t was a P4-type <strong>in</strong>tegrase. <strong>The</strong> P4 family <strong>of</strong> bacteriophages<br />
<strong>in</strong>tegrate downstream <strong>of</strong> tRNA sites and this action is facilitated by <strong>the</strong> <strong>in</strong>tegrase<br />
prote<strong>in</strong>. <strong>The</strong> <strong>in</strong>tegrase is rendered non-functional by two frameshifts. A frameshift is<br />
caused by <strong>in</strong>sertion or deletion <strong>of</strong> nucleotides from a sequence that is not divisible<br />
by 3 (Figure 3-17). <strong>The</strong> earlier <strong>the</strong> disruption with<strong>in</strong> <strong>the</strong> ORF occurs, <strong>the</strong> more