5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
100bp<br />
KS<br />
T3<br />
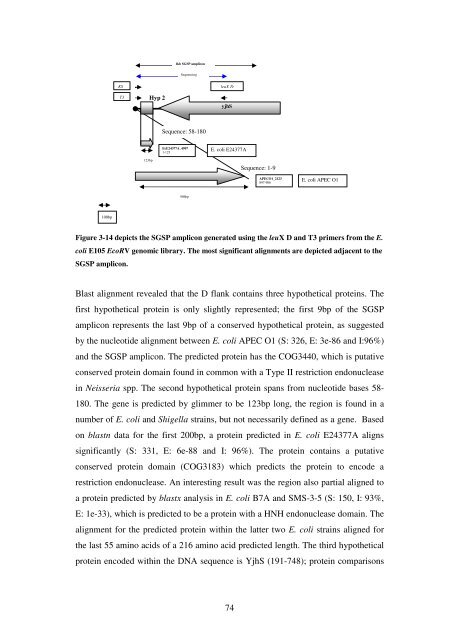
Figure 3-14 depicts <strong>the</strong> SGSP amplicon generated us<strong>in</strong>g <strong>the</strong> leuX D and T3 primers from <strong>the</strong> E.<br />
coli E105 EcoRV genomic library. <strong>The</strong> most significant alignments are depicted adjacent to <strong>the</strong><br />
SGSP amplicon.<br />
123bp<br />
Hyp 2<br />
1kb SGSP amplicon<br />
Sequenc<strong>in</strong>g<br />
Sequence: 58-180<br />
EcE24377A_4907<br />
1-123<br />
906bp<br />
leuX D<br />
yjhS<br />
E. coli E24377A<br />
Blast alignment revealed that <strong>the</strong> D flank conta<strong>in</strong>s three hypo<strong>the</strong>tical prote<strong>in</strong>s. <strong>The</strong><br />
first hypo<strong>the</strong>tical prote<strong>in</strong> is only slightly represented; <strong>the</strong> first 9bp <strong>of</strong> <strong>the</strong> SGSP<br />
amplicon represents <strong>the</strong> last 9bp <strong>of</strong> a conserved hypo<strong>the</strong>tical prote<strong>in</strong>, as suggested<br />
by <strong>the</strong> nucleotide alignment between E. coli APEC O1 (S: 326, E: 3e-86 and I:96%)<br />
and <strong>the</strong> SGSP amplicon. <strong>The</strong> predicted prote<strong>in</strong> has <strong>the</strong> COG3440, which is putative<br />
conserved prote<strong>in</strong> doma<strong>in</strong> found <strong>in</strong> common with a Type II restriction endonuclease<br />
<strong>in</strong> Neisseria spp. <strong>The</strong> second hypo<strong>the</strong>tical prote<strong>in</strong> spans from nucleotide bases 58-<br />
180. <strong>The</strong> gene is predicted by glimmer to be 123bp long, <strong>the</strong> region is found <strong>in</strong> a<br />
number <strong>of</strong> E. coli and Shigella stra<strong>in</strong>s, but not necessarily def<strong>in</strong>ed as a gene. Based<br />
on blastn data for <strong>the</strong> first 200bp, a prote<strong>in</strong> predicted <strong>in</strong> E. coli E24377A aligns<br />
significantly (S: 331, E: 6e-88 and I: 96%). <strong>The</strong> prote<strong>in</strong> conta<strong>in</strong>s a putative<br />
conserved prote<strong>in</strong> doma<strong>in</strong> (COG3183) which predicts <strong>the</strong> prote<strong>in</strong> to encode a<br />
restriction endonuclease. An <strong>in</strong>terest<strong>in</strong>g result was <strong>the</strong> region also partial aligned to<br />
a prote<strong>in</strong> predicted by blastx analysis <strong>in</strong> E. coli B7A and SMS-3-5 (S: 150, I: 93%,<br />
E: 1e-33), which is predicted to be a prote<strong>in</strong> with a HNH endonuclease doma<strong>in</strong>. <strong>The</strong><br />
alignment for <strong>the</strong> predicted prote<strong>in</strong> with<strong>in</strong> <strong>the</strong> latter two E. coli stra<strong>in</strong>s aligned for<br />
<strong>the</strong> last 55 am<strong>in</strong>o acids <strong>of</strong> a 216 am<strong>in</strong>o acid predicted length. <strong>The</strong> third hypo<strong>the</strong>tical<br />
prote<strong>in</strong> encoded with<strong>in</strong> <strong>the</strong> DNA sequence is YjhS (191-748); prote<strong>in</strong> comparisons<br />
74<br />
Sequence: 1-9<br />
APECO1_2123<br />
897-906<br />
E. coli APEC O1