5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
5 The role of quorum-sensing in the virulence of Pseudomonas ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
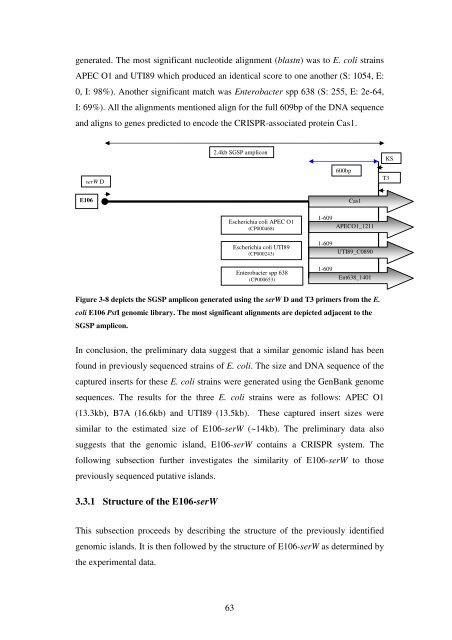
generated. <strong>The</strong> most significant nucleotide alignment (blastn) was to E. coli stra<strong>in</strong>s<br />
APEC O1 and UTI89 which produced an identical score to one ano<strong>the</strong>r (S: 1054, E:<br />
0, I: 98%). Ano<strong>the</strong>r significant match was Enterobacter spp 638 (S: 255, E: 2e-64,<br />
I: 69%). All <strong>the</strong> alignments mentioned align for <strong>the</strong> full 609bp <strong>of</strong> <strong>the</strong> DNA sequence<br />
and aligns to genes predicted to encode <strong>the</strong> CRISPR-associated prote<strong>in</strong> Cas1.<br />
serW D<br />
E106<br />
Figure 3-8 depicts <strong>the</strong> SGSP amplicon generated us<strong>in</strong>g <strong>the</strong> serW D and T3 primers from <strong>the</strong> E.<br />
coli E106 PstI genomic library. <strong>The</strong> most significant alignments are depicted adjacent to <strong>the</strong><br />
SGSP amplicon.<br />
In conclusion, <strong>the</strong> prelim<strong>in</strong>ary data suggest that a similar genomic island has been<br />
found <strong>in</strong> previously sequenced stra<strong>in</strong>s <strong>of</strong> E. coli. <strong>The</strong> size and DNA sequence <strong>of</strong> <strong>the</strong><br />
captured <strong>in</strong>serts for <strong>the</strong>se E. coli stra<strong>in</strong>s were generated us<strong>in</strong>g <strong>the</strong> GenBank genome<br />
sequences. <strong>The</strong> results for <strong>the</strong> three E. coli stra<strong>in</strong>s were as follows: APEC O1<br />
(13.3kb), B7A (16.6kb) and UTI89 (13.5kb). <strong>The</strong>se captured <strong>in</strong>sert sizes were<br />
similar to <strong>the</strong> estimated size <strong>of</strong> E106-serW (~14kb). <strong>The</strong> prelim<strong>in</strong>ary data also<br />
suggests that <strong>the</strong> genomic island, E106-serW conta<strong>in</strong>s a CRISPR system. <strong>The</strong><br />
follow<strong>in</strong>g subsection fur<strong>the</strong>r <strong>in</strong>vestigates <strong>the</strong> similarity <strong>of</strong> E106-serW to those<br />
previously sequenced putative islands.<br />
3.3.1 Structure <strong>of</strong> <strong>the</strong> E106-serW<br />
This subsection proceeds by describ<strong>in</strong>g <strong>the</strong> structure <strong>of</strong> <strong>the</strong> previously identified<br />
genomic islands. It is <strong>the</strong>n followed by <strong>the</strong> structure <strong>of</strong> E106-serW as determ<strong>in</strong>ed by<br />
<strong>the</strong> experimental data.<br />
2.4kb SGSP amplicon<br />
Escherichia coli APEC O1<br />
(CP000468)<br />
63<br />
Escherichia coli UTI89<br />
(CP000243)<br />
Enterobacter spp 638<br />
(CP000653)<br />
1-609<br />
1-609<br />
1-609<br />
600bp<br />
Cas1<br />
APECO1_1211<br />
UTI89_C0890<br />
Ent638_1401<br />
KS<br />
T3