- Page 1 and 2:
Molecular Characterization and Gene
- Page 3 and 4:
Declaration I hereby do declare tha
- Page 5 and 6:
Acknowledgements This work was carr
- Page 7 and 8:
My Seniors, Dr. Annies Joseph, Dr.
- Page 9 and 10:
Contents Title Page Nos. 1 General
- Page 11 and 12:
in response to WSSV challenge 252-2
- Page 13 and 14:
α-2M Alpha-2-Macroglobulin ALF Ant
- Page 15 and 16:
ppA Prophenoloxidase-Activating Enz
- Page 17 and 18:
1.1 Introduction Chapter 1 Aquacult
- Page 19 and 20:
Chapter 1 the giant tiger shrimp (P
- Page 21 and 22:
Common names: Chapter 1 Giant tiger
- Page 23 and 24:
1.3.1. Diseases Chapter 1 Diseases
- Page 25 and 26:
Lymphoidal parvo-like virus (LPV) B
- Page 27 and 28:
Chapter 1 and protective occlusion
- Page 29 and 30:
Chapter 1 small brown masses in the
- Page 31 and 32:
1.4.6. Natural epidemics Chapter 1
- Page 33 and 34:
Chapter 1 can differentiate WSSV-in
- Page 35 and 36:
1.5.1.1. Probiotics Chapter 1 Alter
- Page 37 and 38:
Chapter 1 tried in P. monodon, resu
- Page 39 and 40:
Chapter 1 The first and essential i
- Page 41 and 42:
Chapter 1 1998). The innate immunit
- Page 43 and 44:
1.6.1 Haemocytes Chapter 1 The haem
- Page 45 and 46:
Chapter 1 In crustaceans, the sheet
- Page 47 and 48:
Chapter 1 Theopold et al., 2002). C
- Page 49 and 50:
Chapter 1 kill/remove the invading
- Page 51 and 52:
Chapter 1 forming complexes with a
- Page 53 and 54:
1.7 Antimicrobial Peptides (AMPs) 1
- Page 55 and 56:
Chapter 1 the response is very fast
- Page 57 and 58:
Chapter 1 Hirsch, 1947). While they
- Page 59 and 60:
Chapter 1 AMPs to various degrees u
- Page 61 and 62:
1.7.5 Biological activity Chapter 1
- Page 63 and 64:
Chapter 1 including the microbes as
- Page 65 and 66:
Tachyplesin I Chapter 1 Crustacea P
- Page 67 and 68:
Anionic and cationic peptides that
- Page 69 and 70:
Anionic and Cationic AMPs with cyst
- Page 71 and 72:
Chapter 1 Fig. 1.7. Structural clas
- Page 73 and 74:
Chapter 1 Group III: Linear peptide
- Page 75 and 76:
Chapter 1 through regular processes
- Page 77 and 78:
Chapter 1 ultimately signal to tran
- Page 79 and 80:
Chapter 1 divalent polyanionic cati
- Page 81 and 82:
Chapter 1 type of transmembrane por
- Page 83 and 84:
Chapter 1 other intracellular targe
- Page 85 and 86:
Chapter 1 Apidaecin, a short, proli
- Page 87 and 88:
Chapter 1 Hultmark, 1987; Gennaro a
- Page 89 and 90:
Chapter 1 quite opposite characteri
- Page 91 and 92:
Chapter 1 depletion of mitochondria
- Page 93 and 94:
Chapter 1 conformations adopted by
- Page 95 and 96:
Chapter 1 receptor may be a potenti
- Page 97 and 98:
Chapter 1 exposure to their targets
- Page 99 and 100:
Chapter 1 generating a rich spectru
- Page 101 and 102:
Chapter 1 by enkelytin (Goumon et a
- Page 103 and 104:
Lactoferricin Cow Mastitis control
- Page 105 and 106:
Chapter 1 trial, in which peptide T
- Page 107 and 108:
Chapter 1 a too limited spectrum to
- Page 109 and 110:
Chapter 1 number of research groups
- Page 111 and 112:
CHAPTER-2 Molecular Characterizatio
- Page 113 and 114:
Chapter 2 interaction with negative
- Page 115 and 116:
Chapter 2 inhibit the endotoxin or
- Page 117 and 118:
Chapter 2 structure be modified to
- Page 119 and 120:
Chapter 2 and the region might be l
- Page 121 and 122:
Chapter 2 been made of the spectra
- Page 123 and 124:
Chapter 2 their discovery and initi
- Page 125 and 126:
Chapter 2 terminal pyroglutamic aci
- Page 127 and 128:
Chapter 2 One can assume that the p
- Page 129 and 130:
Chapter 2 by degranulation into the
- Page 131 and 132:
2.2. Materials and Methods 2.2.1. E
- Page 133 and 134:
Chapter 2 washed by adding 1 ml 75
- Page 135 and 136:
Chapter 2 entry of recombinant DNA
- Page 137 and 138:
Chapter 2 and the deduced amino aci
- Page 139 and 140:
Chapter 2 Fenneropenaeus, Litopenae
- Page 141 and 142:
2.3.1.3. Crustin-1 (GQ334395) Chapt
- Page 143 and 144:
2.3.1.3.5. Phylogenetic analysis of
- Page 145 and 146:
Chapter 2 sequence also showed high
- Page 147 and 148:
Chapter 2 chinensis and F. indicus
- Page 149 and 150:
Chapter 2 monodon and Group III of
- Page 151 and 152:
Chapter 2 Tiger shrimp (P. monodon)
- Page 153 and 154:
Chapter 2 ALF-1 was a partial seque
- Page 155 and 156:
Chapter 2 As mentioned before, the
- Page 157 and 158:
Chapter 2 Munoz et al., 2004). The
- Page 159 and 160:
Table 2.1. Primers used for the stu
- Page 161 and 162:
Table 2.4. BLASTn analysis of ALF-2
- Page 163 and 164:
Table 2.8. BLASTn analysis of Crust
- Page 165 and 166:
Table 2.12. BLASTn analysis of Pena
- Page 167 and 168:
Fig.2.5. Agarose gel electrophoreto
- Page 169 and 170:
Chapter 2 Fig.2.9. Multiple alignme
- Page 171 and 172:
Chapter 2 Fig. 2.12 A bootstrapped
- Page 173 and 174:
Chapter 2 Fig. 2.15. Multiple align
- Page 175 and 176:
Chapter 2 Fig.2.18. A bootstrapped
- Page 177 and 178:
Chapter 2 Fig.2.20. Signal peptide
- Page 179 and 180:
Chapter 2 Fig.2.24 Multiple alignme
- Page 181 and 182:
Chapter 2 LITOPENAEUS SP. FARFANTEP
- Page 183 and 184:
Chapter 2 Fig. 2.28. Nucleotide and
- Page 185 and 186:
Chapter 2 Fig.2.33 Multiple alignme
- Page 187 and 188:
Chapter 2 LITOPENAEUS SP. FENNEROPE
- Page 189 and 190:
Chapter 2 Fig. 2.37. Nucleotide and
- Page 191 and 192:
Chapter 2 Fig.2.41 Multiple alignme
- Page 193 and 194:
Chapter 2 Fig.2.43 A bootstrapped n
- Page 195 and 196:
Chapter 2 Fig. 2.45. Nucleotide and
- Page 197 and 198:
Chapter 2 LITOPENAEUS SP. FENNEROPE
- Page 199 and 200:
Chapter 2 Fig.2.51 Signal peptide a
- Page 201 and 202:
Chapter 2 FENNEROPENAEUS SP. FARFAN
- Page 203 and 204:
CHAPTER-3 Molecular Characterizatio
- Page 205 and 206:
Chapter 3 certainly help us to unra
- Page 207 and 208:
Chapter 3 3.3.1.1. Anti-lipopolysac
- Page 209 and 210:
Chapter 3 was predicted out to be 1
- Page 211 and 212:
Chapter 3 random coiled structure t
- Page 213 and 214:
3.3.1.3.5. Phylogenetic analysis of
- Page 215 and 216:
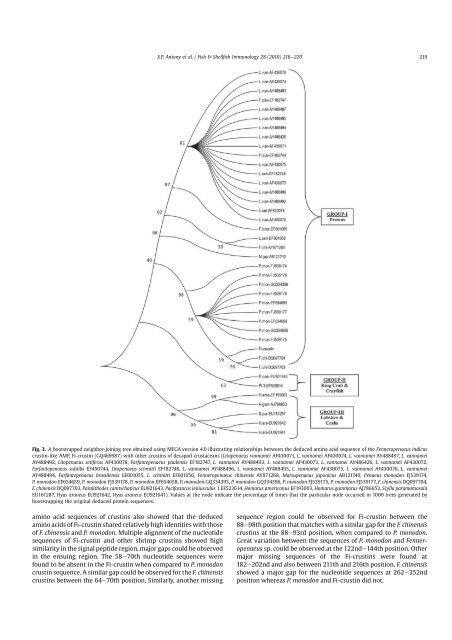
Chapter 3 case has been reported in
- Page 217 and 218:
Chapter 3 Multiple alignments were
- Page 219 and 220:
Chapter 3 negative bacteria. In shr
- Page 221 and 222:
Chapter 3 indicated a random coiled
- Page 223 and 224:
Chapter 3 horseshoe crab big defens
- Page 225 and 226:
Table 3.4. BLASTn analysis of ALF-2
- Page 227 and 228:
Table.3.8. BLASTn analysis of Fi-pe
- Page 229 and 230:
Chapter 3 Fig.3.2. Nucleotide and a
- Page 231 and 232:
Chapter 3 Fig.3.5. Multiple alignme
- Page 233 and 234:
Chapter 3 Fig.3.8. Nucleotide and a
- Page 235 and 236:
Chapter 3 Fig.3.12. Multiple alignm
- Page 237 and 238:
Chapter 3 Fig.3.14. A bootstrapped
- Page 239 and 240:
Chapter 3 Fig.3.17. Signal peptide
- Page 241 and 242:
Chapter 3 Fig.3.21. Multiple alignm
- Page 243 and 244:
SHRIMPS KIND CRAB LOBSTERS CRABS Ch
- Page 245 and 246:
Chapter 3 Fig.3.25. Signal peptide
- Page 247 and 248:
Chapter 3 Fig. 3.28. Multiple align
- Page 249 and 250:
PENAEIDIN-2 Chapter 3 PENAEIDIN- 3
- Page 251 and 252:
Fig.3.31. Nucleotide and amino acid
- Page 253 and 254:
Chapter 3 Fig.3.35. Multiple alignm
- Page 255 and 256:
Chapter 3 Fig.3.37 Nucleotide and a
- Page 257 and 258:
4.1. Introduction Chapter 4 The aqu
- Page 259 and 260:
Chapter 4 appearance and have major
- Page 261 and 262:
Chapter 4 the tissues has recently
- Page 263 and 264:
Chapter 4 with the ability of shrim
- Page 265 and 266:
Chapter 4 with a low dose of WSSV b
- Page 267 and 268:
Chapter 4 molecules they are likely
- Page 269 and 270:
Chapter 4 for understanding gene ex
- Page 271 and 272:
Chapter 4 4.3.2. Expression profile
- Page 273 and 274:
Expression profile of endonuclease
- Page 275 and 276:
Chapter 4 bacterial and fungal infe
- Page 277 and 278:
Chapter 4 measuring TSV and YHV loa
- Page 279 and 280:
Chapter 4 Of particular interest he
- Page 281 and 282:
Chapter 4 large distribution and ab
- Page 283 and 284:
Chapter 4 WSSV genes viz. endonucle
- Page 285 and 286:
Table 4.2. Primers used for the stu
- Page 287 and 288:
(A) (B) B 28 1 2 3 4 5 6 7 8 9 10 C
- Page 289 and 290:
(A) (B) B 28 1 2 3 4 5 6 7 8 9 10 C
- Page 291 and 292:
(A) (B) B 28 1 2 3 4 5 6 7 8 9 10 C
- Page 293 and 294:
(A) (B) B 28 1 2 3 4 5 6 7 8 9 10 C
- Page 295 and 296:
(A) (B) B 28 1 2 3 4 5 6 7 8 9 10 F
- Page 297 and 298:
(A) (B) B 28 1 2 3 4 5 6 7 8 9 10 C
- Page 299 and 300:
(A) (B) B 28 1 2 3 4 5 6 7 8 9 10 C
- Page 301 and 302:
(A) (B) B 28 1 2 3 4 5 6 7 8 9 10 C
- Page 303 and 304:
CHAPTER-5 Expression Profile of Ant
- Page 305 and 306:
Chapter 5 1994). The emergence of b
- Page 307 and 308:
Chapter 5 (Bovill et al., 2001). In
- Page 309 and 310:
5.2.2. Test diets Chapter 5 Four ex
- Page 311 and 312:
5.2.2.4. Preparation of Glucan inco
- Page 313 and 314:
Chapter 5 genes (latency related ge
- Page 315 and 316:
Chapter 5 On WSSV challenge, crusti
- Page 317 and 318:
Chapter 5 5.3.4. Expression profile
- Page 319 and 320:
5.3.4.5. Expression profile of pena
- Page 321 and 322:
Chapter 5 must firstly rest on a so
- Page 323 and 324:
Chapter 5 For the present study two
- Page 325 and 326:
Chapter 5 ameobocytes of two marine
- Page 327 and 328:
Chapter 5 variation in expression o
- Page 329 and 330:
Chapter 5 It has been previously sh
- Page 331 and 332:
Chapter 5 manifested in terms of gr
- Page 333 and 334:
(A) (B) (C) C CHY CSY CHG CSG C CHY
- Page 335 and 336:
(A) (B) (C) C CHY CSY CHG CSG C CHY
- Page 337 and 338:
(A) (B) (C) C CHY CSY CHG CSG C CHY
- Page 339 and 340:
(A) (B) (C) C CHY CSY CHG CSG C CHY
- Page 341 and 342:
(A) (B) -VE CTRL +VE CTRL CHG CSG C
- Page 343 and 344:
(A) (B) -VE CTRL +VE CTRL CHG CSG C
- Page 345 and 346:
(A) (B) -VE CTRL +VE CTRL CHG CSG C
- Page 347 and 348:
(A) (B) -VE CTRL +VE CTRL CHG CSG C
- Page 349 and 350:
(A) PRE-CHALLENGE (B) POST-CHALLENG
- Page 351 and 352:
(A) PRE-CHALLENGE (B) (C) (D) CONTR
- Page 353 and 354:
(C) (D) (A) PRE-CHALLENGE (B) CONTR
- Page 355 and 356:
(A) (B) G M HP HR I Chapter 5 Fig.
- Page 357 and 358:
Chapter 5 Fig. 5.26. Post challenge
- Page 359 and 360:
6.1. Introduction Chapter 6 Prophyl
- Page 361 and 362:
Chapter 6 production of inhibitory
- Page 363 and 364:
6.2. Materials and methods 6.2.1. E
- Page 365 and 366:
Chapter 6 tissue cDNA was performed
- Page 367 and 368:
Chapter 6 gene was supported by Bac
- Page 369 and 370:
Chapter 6 treated group of shrimps.
- Page 371 and 372:
Chapter 6 the gene was found to be
- Page 373 and 374:
Chapter 6 found to support minimum
- Page 375 and 376:
Chapter 6 is induced upon bacterial
- Page 377 and 378:
Chapter 6 all the target genes, exc
- Page 379 and 380:
Chapter 6 Detailed tissue-wise anal
- Page 381 and 382:
Chapter 6 Gills and intestine was f
- Page 383 and 384:
(A) (B) Fig. 6.1. Experimental diet
- Page 385 and 386:
(A) (B) (C) Fig. 6.3. Expression pr
- Page 387 and 388:
(A) (B) (C) C B M BM C B M BM Fig.
- Page 389 and 390:
(A) (B) (C) Fig. 6.7. Expression pr
- Page 391 and 392:
(A) (B) (C) Chapter 6 Fig. 6.9. Exp
- Page 393 and 394:
(A) (B) -VE CTRL +VE CTRL B M BM Ch
- Page 395 and 396:
(A) (B) -VE CTRL +VE CTRL B M BM Ch
- Page 397 and 398:
(A) (B) -VE CTRL +VE CTRL B M BM Ch
- Page 399 and 400:
(A) (B) -VE CTRL +VE CTRL B M BM Ch
- Page 401 and 402:
(A) PRE-CHALLENGE (B) POST-CHALLENG
- Page 403 and 404:
(A) PRE-CHALLENGE (B) POST-CHALLENG
- Page 405 and 406:
(A) (C) (D) PRE-CHALLENGE POST-CHAL
- Page 407 and 408:
(A) (B) G M HP HR I Chapter 6 Fig.
- Page 409 and 410:
Chapter 6 Fig. 6.27. Post challenge
- Page 411 and 412:
7.1. Summary Chapter 7 Tropical shr
- Page 413 and 414:
Salient findings Chapter 7 � From
- Page 415 and 416:
Chapter 7 � Tissue-wise expressio
- Page 417 and 418:
Chapter 7 application in shrimp cul
- Page 419 and 420:
References Aboudy, Y., Mendelson, E
- Page 421 and 422:
References Anggraeni, M.S., Owens,
- Page 423 and 424:
References Bals, R., Wang, X., Zasl
- Page 425 and 426:
References derived from the N-termi
- Page 427 and 428:
References Breukink, E., de Kruijff
- Page 429 and 430:
References Burgents, J.E., Burnett,
- Page 431 and 432:
References 1,3-β-D-glucan-binding
- Page 433 and 434:
References Chen, J.Y., Chuang, H.,
- Page 435 and 436:
References Cole, A.M., Hong, T., Bo
- Page 437 and 438:
References Dathe, M., Wieprecht, T.
- Page 439 and 440:
References Diamond, G., Zasloff, M.
- Page 441 and 442:
References Faber, C., Stallmann, H.
- Page 443 and 444:
References Flint, S.J., Enquist, L.
- Page 445 and 446:
References Gennaro, R., Zanetti, M.
- Page 447 and 448:
References Gudmundsson, G.H., Lidho
- Page 449 and 450:
References Harwig, S.S., Swiderek,
- Page 451 and 452:
References Hirakura, Y., Kobayashi,
- Page 453 and 454:
References Huang, C.C., Song, Y.L.
- Page 455 and 456:
References Itami, T., Takahashi, Y.
- Page 457 and 458:
References Jiravanichpaisal, P. Whi
- Page 459 and 460:
References Kang, J.H., Shin, S.Y.,
- Page 461 and 462:
References Kono, T., Savan, R., Sak
- Page 463 and 464:
References rich peptide derived fro
- Page 465 and 466:
References Li, L., Wang, J.X., Zhao
- Page 467 and 468:
References Liu, C.H., Cheng, W., Ku
- Page 469 and 470:
References Lorenzon, S., de Guarrin
- Page 471 and 472:
References Marone, M., Mozzetti, S.
- Page 473 and 474:
References Medvinsky, A., Dzierzak,
- Page 475 and 476:
References Moriarty, D.J.W. 1996. M
- Page 477 and 478:
References antimicrobial peptide fr
- Page 479 and 480:
References Otta, S.K., Karunasagar,
- Page 481 and 482:
References simplex virus has heptad
- Page 483 and 484:
References infected shrimp. Thesis
- Page 485 and 486:
References Litopenaeus vannamei cha
- Page 487 and 488:
References Roth, B.L., Poot, M., Yu
- Page 489 and 490: References Sathish, S., Musthaq, S.
- Page 491 and 492: References a proline-arginine-rich
- Page 493 and 494: References Smith, V.J., Fernandes,
- Page 495 and 496: References Song, Y.L., Cheng, W., W
- Page 497 and 498: References experimental system. In:
- Page 499 and 500: References the black tiger shrimp P
- Page 501 and 502: References Tapay, L.M., Cesar, E.,
- Page 503 and 504: References Touhy, K.M., Probert, H.
- Page 505 and 506: References van Hulten, M.C., Reijns
- Page 507 and 508: References Vives, R.R., Imberty, A.
- Page 509 and 510: References Wang, Y.C., Chang, P.S.,
- Page 511 and 512: References Williams, M.J., Rodrigue
- Page 513 and 514: References Wu, W., Zong, R., Xu, J.
- Page 515 and 516: References Yodmuang, S., Tirasophon
- Page 517 and 518: References Zhan, W.B., Wang, Y.H. 1
- Page 519 and 520: Penaeus monodon anti-lipopolysaccha
- Page 521 and 522: Penaeus monodon crustin-like antimi
- Page 523 and 524: Penaeus monodon crustin-like antimi
- Page 525 and 526: Penaeus monodon penaeidin-like anti
- Page 527 and 528: Penaeus monodon elongation factor m
- Page 529 and 530: Fenneropenaeus indicus anti-lipopol
- Page 531 and 532: Fenneropenaeus indicus penaeidin-li
- Page 533 and 534: Fenneropenaeus indicus beta-actin m
- Page 535 and 536: PUBLICATIONS
- Page 537 and 538: Short sequence report Molecular cha
- Page 539: Fig. 2. Multiple alignment of nucle
- Page 543 and 544: Table 1 Rearing conditions and wate
- Page 545 and 546: 5 ′ cttgtggttgacaatggctccg3
- Page 547 and 548: Expression of WSSV genes in the hae
- Page 549 and 550: Fig. 4. Expression profile of crust
- Page 551 and 552: S.P. Antony et al. / Immunobiology