Thesis Title: Subtitle - NMR Spectroscopy Research Group
Thesis Title: Subtitle - NMR Spectroscopy Research Group
Thesis Title: Subtitle - NMR Spectroscopy Research Group
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
106 Chapter 4. Protein Structure Determination from Pseudocontact Shifts using ROSETTA.<br />
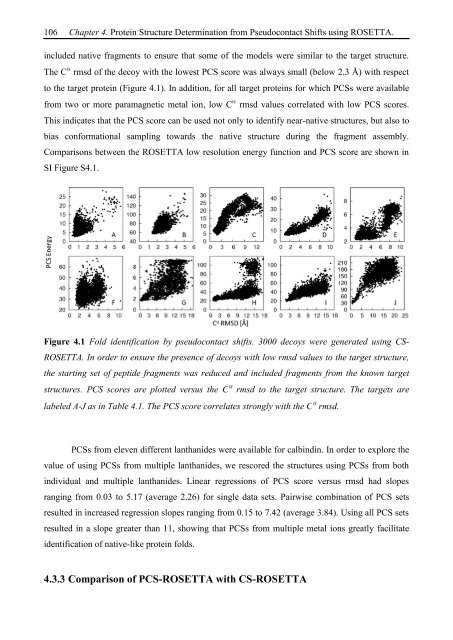
included native fragments to ensure that some of the models were similar to the target structure.<br />
The C rmsd of the decoy with the lowest PCS score was always small (below 2.3 Å) with respect<br />
to the target protein (Figure 4.1). In addition, for all target proteins for which PCSs were available<br />
from two or more paramagnetic metal ion, low C rmsd values correlated with low PCS scores.<br />
This indicates that the PCS score can be used not only to identify near-native structures, but also to<br />
bias conformational sampling towards the native structure during the fragment assembly.<br />
Comparisons between the ROSETTA low resolution energy function and PCS score are shown in<br />
SI Figure S4.1.<br />
Figure 4.1 Fold identification by pseudocontact shifts. 3000 decoys were generated using CS-<br />
ROSETTA. In order to ensure the presence of decoys with low rmsd values to the target structure,<br />
the starting set of peptide fragments was reduced and included fragments from the known target<br />
structures. PCS scores are plotted versus the C rmsd to the target structure. The targets are<br />
labeled A-J as in Table 4.1. The PCS score correlates strongly with the C rmsd.<br />
PCSs from eleven different lanthanides were available for calbindin. In order to explore the<br />
value of using PCSs from multiple lanthanides, we rescored the structures using PCSs from both<br />
individual and multiple lanthanides. Linear regressions of PCS score versus rmsd had slopes<br />
ranging from 0.03 to 5.17 (average 2.26) for single data sets. Pairwise combination of PCS sets<br />
resulted in increased regression slopes ranging from 0.15 to 7.42 (average 3.84). Using all PCS sets<br />
resulted in a slope greater than 11, showing that PCSs from multiple metal ions greatly facilitate<br />
identification of native-like protein folds.<br />
4.3.3 Comparison of PCS-ROSETTA with CS-ROSETTA