Computational tools and Interoperability in Comparative ... - CBS
Computational tools and Interoperability in Comparative ... - CBS
Computational tools and Interoperability in Comparative ... - CBS
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
2 O. N. Reva et al.<br />
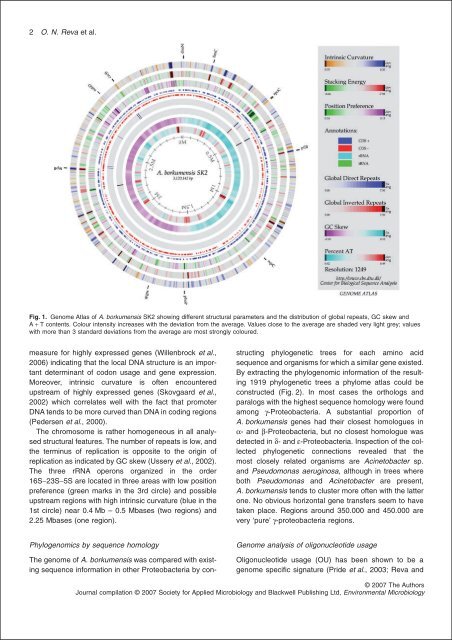
Fig. 1. Genome Atlas of A. borkumensis SK2 show<strong>in</strong>g different structural parameters <strong>and</strong> the distribution of global repeats, GC skew <strong>and</strong><br />
A + T contents. Colour <strong>in</strong>tensity <strong>in</strong>creases with the deviation from the average. Values close to the average are shaded very light grey; values<br />
with more than 3 st<strong>and</strong>ard deviations from the average are most strongly coloured.<br />
measure for highly expressed genes (Willenbrock et al.,<br />
2006) <strong>in</strong>dicat<strong>in</strong>g that the local DNA structure is an important<br />
determ<strong>in</strong>ant of codon usage <strong>and</strong> gene expression.<br />
Moreover, <strong>in</strong>tr<strong>in</strong>sic curvature is often encountered<br />
upstream of highly expressed genes (Skovgaard et al.,<br />
2002) which correlates well with the fact that promoter<br />
DNA tends to be more curved than DNA <strong>in</strong> cod<strong>in</strong>g regions<br />
(Pedersen et al., 2000).<br />
The chromosome is rather homogeneous <strong>in</strong> all analysed<br />
structural features. The number of repeats is low, <strong>and</strong><br />
the term<strong>in</strong>us of replication is opposite to the orig<strong>in</strong> of<br />
replication as <strong>in</strong>dicated by GC skew (Ussery et al., 2002).<br />
The three rRNA operons organized <strong>in</strong> the order<br />
16S-23S-5S are located <strong>in</strong> three areas with low position<br />
preference (green marks <strong>in</strong> the 3rd circle) <strong>and</strong> possible<br />
upstream regions with high <strong>in</strong>tr<strong>in</strong>sic curvature (blue <strong>in</strong> the<br />
1st circle) near 0.4 Mb – 0.5 Mbases (two regions) <strong>and</strong><br />
2.25 Mbases (one region).<br />
Phylogenomics by sequence homology<br />
The genome of A. borkumensis was compared with exist<strong>in</strong>g<br />
sequence <strong>in</strong>formation <strong>in</strong> other Proteobacteria by con-<br />
struct<strong>in</strong>g phylogenetic trees for each am<strong>in</strong>o acid<br />
sequence <strong>and</strong> organisms for which a similar gene existed.<br />
By extract<strong>in</strong>g the phylogenomic <strong>in</strong>formation of the result<strong>in</strong>g<br />
1919 phylogenetic trees a phylome atlas could be<br />
constructed (Fig. 2). In most cases the orthologs <strong>and</strong><br />
paralogs with the highest sequence homology were found<br />
among g-Proteobacteria. A substantial proportion of<br />
A. borkumensis genes had their closest homologues <strong>in</strong><br />
a- <strong>and</strong> b-Proteobacteria, but no closest homologue was<br />
detected <strong>in</strong> d- <strong>and</strong> e-Proteobacteria. Inspection of the collected<br />
phylogenetic connections revealed that the<br />
most closely related organisms are Ac<strong>in</strong>etobacter sp.<br />
<strong>and</strong> Pseudomonas aerug<strong>in</strong>osa, although <strong>in</strong> trees where<br />
both Pseudomonas <strong>and</strong> Ac<strong>in</strong>etobacter are present,<br />
A. borkumensis tends to cluster more often with the latter<br />
one. No obvious horizontal gene transfers seem to have<br />
taken place. Regions around 350.000 <strong>and</strong> 450.000 are<br />
very ‘pure’ g-proteobacteria regions.<br />
Genome analysis of oligonucleotide usage<br />
Oligonucleotide usage (OU) has been shown to be a<br />
genome specific signature (Pride et al., 2003; Reva <strong>and</strong><br />
©2007TheAuthors<br />
Journal compilation © 2007 Society for Applied Microbiology <strong>and</strong> Blackwell Publish<strong>in</strong>g Ltd, Environmental Microbiology