THIS WEEK IN
THIS WEEK IN
THIS WEEK IN
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
R EPORTS<br />
We analyzed the intracellular localization of<br />
the miRNP components. Previous work has indicated<br />
that translationally repressed mRNAs<br />
can be sequestered in specific cellular structures<br />
such as germline (13) or stress (14) granules.<br />
Immunofluorescence analysis indicated that<br />
HA-tagged hAgo2-4 proteins, expressed in transfected<br />
HeLa and human embryonic kidney<br />
(HEK) 293 cells (fig. S7) or in stable HeLa cell<br />
lines (figs. S8, A to C, and S9A), localize to<br />
processing bodies (PBs; visualized as structures<br />
enriched in the marker proteins Lsm1, Dcp1a,<br />
and Xrn1), suborganelles identified as sites of<br />
mRNA degradation (15). Association of Ago<br />
proteins with PBs was further confirmed by<br />
co-immunoprecipitation experiments (fig. S8,<br />
D and E). Notably, the GFP-tagged Dcp1<br />
was found to interact with HA-hAgo2 but not<br />
HA-hAgo2 DPRP , a mutant of hAgo2 that does<br />
not function as a repressor (8) (fig. S8E).<br />
Next, we analyzed the cellular distribution<br />
of reporter mRNAs by in situ hybridization in<br />
HeLa cells. The RL-3xBulge mRNA but not its<br />
mutant version, RL-3xBulgeMut, co-localized<br />
with the PB marker Dcp1a, expressed as a fusion<br />
with green fluorescent protein (GFP) (Fig.<br />
3A and table S1). The co-localization of RL-<br />
3xBulge mRNA with PBs was let-7–dependent<br />
(table S2). Moreover, RL-3xBulgeMut RNA<br />
A<br />
RL (arbitrary units)<br />
7<br />
6<br />
5<br />
4<br />
3<br />
2<br />
1<br />
0<br />
RL<br />
Con<br />
C<br />
FL (normalized)<br />
2.5<br />
2<br />
1.5<br />
1<br />
0.5<br />
0<br />
Perf<br />
NHA-4E<br />
Con<br />
1xBulge<br />
+<br />
P olyA<br />
-<br />
PolyA<br />
3xBulge<br />
FL<br />
NHA-4G<br />
3xBulge<br />
NHA-LacZ<br />
B<br />
FL or RL activities<br />
(normalized)<br />
2 BoxB<br />
accumulated in PBs when co-transfected with<br />
the siRNA-like duplex, let-7Mut, which carries<br />
mutations restoring base pairing between let-7<br />
and RL-3xBulgeMut RNAs (Fig. 3A and fig.<br />
S10). The RL-Perf mRNA could not be<br />
visualized in PBs, arguing against a possibility<br />
that RL-3xBulge or RL-3xBulgeMut signals in<br />
PBs originate from an mRNA targeted for degradation<br />
(fig. S11). Closer examination of the<br />
images provided additional evidence that RL-<br />
3xBulge RNA localization is not related to the<br />
degradative function of PBs. RL-3xBulge RNA<br />
was often found adjacent to Dcp1 foci and not<br />
overlapping with them (Fig. 3A, fig. S11, and<br />
table S3). Furthermore, we also observed that<br />
some Dcp1 foci did not contain detectable<br />
amounts of RL-3xBulge RNA and, conversely,<br />
that a few mRNA foci were negative for Dcp1<br />
(9). These data point to some heterogeneity<br />
and/or compartmentalization of PBs.<br />
We used HeLa cells stably expressing either<br />
RL-3xBulge or RL-3xBulgeMut reporters<br />
(fig. S9, B and C) to further document the association<br />
of repressed mRNAs with cellular<br />
structures. The cells were permeabilized with<br />
digitonine, and the resulting S14 supernatant and<br />
pellet fractions were analyzed for the presence<br />
of mRNAs and PB and miRNP components.<br />
The RL-3xBulgeMut RNA was enriched in a<br />
RL (normalized)<br />
1.5<br />
1<br />
0.5<br />
0<br />
1.5<br />
1<br />
0.5<br />
0<br />
Con<br />
FL<br />
Con<br />
3xBulge<br />
RL<br />
NHA-4E<br />
Con<br />
EMCV-FL<br />
3xBulge<br />
Con<br />
EMCV-RL<br />
3xBulge<br />
Anti-miR-122a<br />
Anti-let-7<br />
NHA-4G<br />
Con or<br />
3xBulge<br />
3xBulge<br />
NHA-LacZ<br />
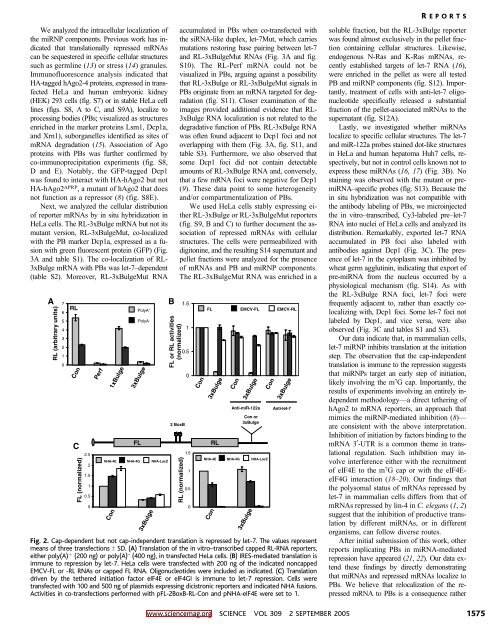
Fig. 2. Cap-dependent but not cap-independent translation is repressed by let-7. The values represent<br />
means of three transfections T SD. (A) Translation of the in vitro–transcribed capped RL-RNA reporters,<br />
either poly(A) þ (200 ng) or poly(A) – (400 ng), in transfected HeLa cells. (B) IRES-mediated translation is<br />
immune to repression by let-7. HeLa cells were transfected with 200 ng of the indicated noncapped<br />
EMCV-FL or -RL RNAs or capped FL RNA. Oligonucleotides were included as indicated. (C) Translation<br />
driven by the tethered initiation factor eIF4E or eIF4GI is immune to let-7 repression. Cells were<br />
transfected with 100 and 500 ng of plasmids expressing dicistronic reporters and indicated NHA fusions.<br />
Activities in co-transfections performed with pFL-2BoxB-RL-Con and pNHA-eIF4E were set to 1.<br />
soluble fraction, but the RL-3xBulge reporter<br />
was found almost exclusively in the pellet fraction<br />
containing cellular structures. Likewise,<br />
endogenous N-Ras and K-Ras mRNAs, recently<br />
established targets of let-7 RNA (16),<br />
were enriched in the pellet as were all tested<br />
PB and miRNP components (fig. S12). Importantly,<br />
treatment of cells with anti-let-7 oligonucleotide<br />
specifically released a substantial<br />
fraction of the pellet-associated mRNAs to the<br />
supernatant (fig. S12A).<br />
Lastly, we investigated whether miRNAs<br />
localize to specific cellular structures. The let-7<br />
and miR-122a probes stained dot-like structures<br />
in HeLa and human hepatoma Huh7 cells, respectively,<br />
but not in control cells known not to<br />
express these miRNAs (16, 17) (Fig. 3B). No<br />
staining was observed with the mutant or premiRNA–specific<br />
probes (fig. S13). Because the<br />
in situ hybridization was not compatible with<br />
the antibody labeling of PBs, we microinjected<br />
the in vitro–transcribed, Cy3-labeled pre–let-7<br />
RNA into nuclei of HeLa cells and analyzed its<br />
distribution. Remarkably, exported let-7 RNA<br />
accumulated in PB foci also labeled with<br />
antibodies against Dcp1 (Fig. 3C). The presence<br />
of let-7 in the cytoplasm was inhibited by<br />
wheat germ agglutinin, indicating that export of<br />
pre-miRNA from the nucleus occurred by a<br />
physiological mechanism (fig. S14). As with<br />
the RL-3xBulge RNA foci, let-7 foci were<br />
frequently adjacent to, rather than exactly colocalizing<br />
with, Dcp1 foci. Some let-7 foci not<br />
labeled by Dcp1, and vice versa, were also<br />
observed (Fig. 3C and tables S1 and S3).<br />
Our data indicate that, in mammalian cells,<br />
let-7 miRNP inhibits translation at the initiation<br />
step. The observation that the cap-independent<br />
translation is immune to the repression suggests<br />
that miRNPs target an early step of initiation,<br />
likely involving the m 7 G cap. Importantly, the<br />
results of experiments involving an entirely independent<br />
methodology—a direct tethering of<br />
hAgo2 to mRNA reporters, an approach that<br />
mimics the miRNP-mediated inhibition (8)—<br />
are consistent with the above interpretation.<br />
Inhibition of initiation by factors binding to the<br />
mRNA 3-UTR is a common theme in translational<br />
regulation. Such inhibition may involve<br />
interference either with the recruitment<br />
of eIF4E to the m 7 G cap or with the eIF4EeIF4G<br />
interaction (18–20). Our findings that<br />
the polysomal status of mRNAs repressed by<br />
let-7 in mammalian cells differs from that of<br />
mRNAs repressed by lin-4 in C. elegans (1, 2)<br />
suggest that the inhibition of productive translation<br />
by different miRNAs, or in different<br />
organisms, can follow diverse routes.<br />
After initial submission of this work, other<br />
reports implicating PBs in miRNA-mediated<br />
repression have appeared (21, 22). Our data extend<br />
these findings by directly demonstrating<br />
that miRNAs and repressed mRNAs localize to<br />
PBs. We believe that relocalization of the repressed<br />
mRNA to PBs is a consequence rather<br />
www.sciencemag.org SCIENCE VOL 309 2 SEPTEMBER 2005 1575