THIS WEEK IN
THIS WEEK IN
THIS WEEK IN
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
R EPORTS<br />
spliceosomal introns (1)^. Thus, the evolution<br />
of GIR1 ribozymes illustrates a possible<br />
step in the evolution of spliceosomal introns.<br />
The homing endonuclease is expressed from<br />
I-DirI mRNA formed by precursor ribosomal<br />
RNA (rRNA) processing (10). The formation<br />
of the 5 end of the I-DirI mRNAbycleavage<br />
of the spliced out intron precludes the normal<br />
cotranscriptional addition of a cap nucleotide.<br />
The cap nucleotide in typical messengers serves<br />
several functions, including protection from<br />
degradation by 5Y3 exonucleases, and recruitment<br />
of protein factors in initiation of<br />
translation (15). The reaction at IPS in GIR1<br />
results in protection of the 5 end of the I-DirI<br />
mRNA against the activity of 5Y3 exonucleases<br />
by formation of a lariat cap. This<br />
potentially substitutes for one of the features<br />
of the missing cap nucleotide. GIR1 processed<br />
RNAs are stable when expressed in Escherichia<br />
coli and yeast (11, 16), which supports this idea<br />
of 5 stabilization by lariat formation.<br />
The present observations extend the diversity<br />
of natural ribozymes (3) and connect the group I<br />
and group II introns, two major structural<br />
classes of self-splicing RNAs. Despite its<br />
overall structural similarity with other natural<br />
group I introns, the GIR1 ribozyme is distinct<br />
from the group I splicing ribozymes not only in<br />
key structural features but also in the type of<br />
reaction catalyzed and in the derived function.<br />
References and Notes<br />
1. C. B. Burge, T. Tuschl, P. A. Sharp, in The RNA<br />
World, Second Edition, R.F.Gesteland,T.R.Cech,<br />
J. F. Atkins, Eds. [Cold Spring Harbor Laboratory<br />
(CSHL) Press, Cold Spring Harbor, New York,<br />
1999], pp. 525–560.<br />
2. C. R. Trotta, J. Abelson, in The RNA World, Second<br />
Edition, R. F. Gesteland, T. R. Cech, J. F. Atkins, Eds.<br />
(CSHL Press, Cold Spring Harbor, New York, 1999),<br />
pp. 561–584.<br />
Fig. 2. (A) Characterization of the 5 end of the 22-nt 3 fragment. [ 32 P]pCplabeled<br />
3 fragment was isolated from 157.22 and 166.22 and subjected to<br />
treatment with alkaline phosphatase (AP), AP followed by rephosphorylation<br />
with T4 polynucleotide kinase (APxPNK), or treatment with PNK alone (PNK).<br />
The sample denoted (157)22 166 was preincubated at reaction conditions<br />
for 30 min before the analysis. OH and T1: Alkaline ladder and T1 digest of<br />
[ 32 P]pCp–labeled precursor 157.22. (B) Diagram of the 22 nt lariat used for<br />
experiments in (C) and (D). The RNA was body-labeled at the phosphates in<br />
bold by incorporation of 32 P. Arrows indicate potential cleavage sites for mung<br />
bean nuclease (MB) and snake venom phosphodiesterase (SV). Cleavage of the<br />
22-nt fragment at sites labeled 1 with SV results in a protected lariat circle<br />
(LC). Cleavage at sites labeled 1 and 2 with MB results in a protected branched<br />
nucleotide (BR). Subsequent cleavage of BR with SV at sites labeled 3 releases<br />
the nucleotides involved in the branch. (C) Characterization of the lariat circle<br />
by gel purification and subsequent digestion with MB (LCxMB). The 22-nt<br />
fragment and digests with MB or SV serves as markers. (D) Characterization of<br />
the branched nucleotide by purification of its phosphorylated and dephosphorylated<br />
form, and subsequent TLC analysis of nucleotides liberated by<br />
digestion with SV. The first two runs show digests of the 22-nt fragment. The<br />
followingshowtheisolatedbranch(BR),anddephosphorylatedbranch(BRAP),<br />
respectively. Finally, the last two runs show the subsequent digests of these<br />
with SV (BRxSV and BRAPxSV).<br />
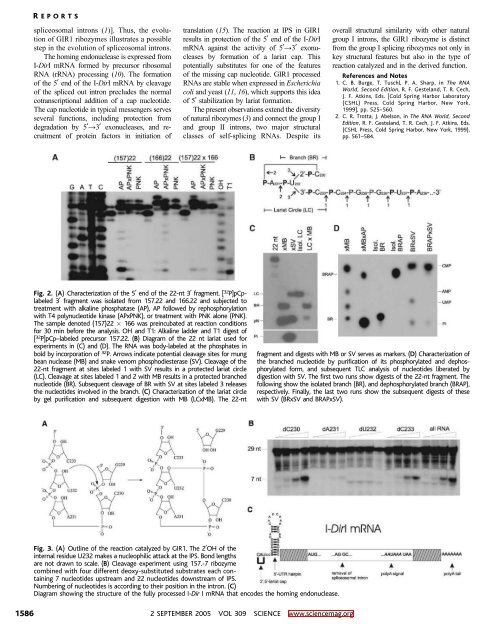
Fig. 3. (A) Outline of the reaction catalyzed by GIR1. The 2OH of the<br />
internal residue U232 makes a nucleophilic attack at the IPS. Bond lengths<br />
are not drawn to scale. (B) Cleavage experiment using 157.-7 ribozyme<br />
combined with four different deoxy-substituted substrates each containing<br />
7 nucleotides upstream and 22 nucleotides downstream of IPS.<br />
Numbering of nucleotides is according to their position in the intron. (C)<br />
Diagram showing the structure of the fully processed I-Dir I mRNA that encodes the homing endonuclease.<br />
1586<br />
2 SEPTEMBER 2005 VOL 309 SCIENCE www.sciencemag.org