THIS WEEK IN
THIS WEEK IN
THIS WEEK IN
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
R EPORTS<br />
tion E(157)22 RNA^ with AP (alkaline phosphatase)<br />
or AP and PNK (polynucleotide kinase),<br />
or PNK alone all shifted the mobility of the<br />
fragment one position upward in the gel, which<br />
was consistent with the removal of the 3-<br />
phosphate of the pCp label. In contrast, a 3<br />
fragment that resulted from cleavage at IPS1<br />
without the IPS2 modification E(166)22 RNA^<br />
was shifted two positions upward with AP, one<br />
position when phosphorylated with PNK after<br />
AP treatment, and one position with PNK<br />
alone. This is consistent with removal of the<br />
3-phosphate (from the pCp) as well as an additional<br />
phosphate at the 5 endleftbyIPS1<br />
cleavage. Thus, the phosphate at the 5 end of<br />
the 22-nt 3 fragment is accessible to phosphatase<br />
in the absence of the IPS2 modification<br />
but inaccessible when the IPS2 modification is<br />
present. This feature of the IPS2 modification<br />
could be removed by incubation of (157)22<br />
RNA with 166 RNA before the analysis, as<br />
shown in the last panel in Fig. 2A. Thus, both<br />
the primer extension stop at IPS2 and blocking<br />
of the 5 end are reversible. An explanation<br />
for these observations is that GIR1<br />
cleavage occurs by a transesterification reaction<br />
in which cleavage at IPS1 is coupled to<br />
formation of a 2,5-phosphodiester bond between<br />
C230 and U232. This explains the<br />
primer extension stop at IPS2, the blocking of<br />
the 5 end, the conservation of internal energy<br />
after cleavage, and the reversibility of the reaction.<br />
Consistent with the engagement of the<br />
2OH of U232, this nucleotide is resistant to<br />
alkaline hydrolysis in (157)22 RNA, in contrast<br />
to (166)22 RNA (fig. S3).<br />
Branches in RNA are resistant to digestion<br />
with various RNases including mung bean<br />
nuclease (13). A resistant fragment was found<br />
in mung bean nuclease digests of bodylabeled<br />
(157)22 RNA but not (166)22 RNA<br />
(fig. S4 and SOM text). Digestion of (157)22<br />
RNA with the exonuclease snake venom<br />
phosphodiesterase resulted in a resistant fragment<br />
corresponding to the 4-nt lariat circle<br />
(Fig. 2B) that could subsequently be cleaved<br />
by the endonuclease mung bean nuclease to<br />
release the branched nucleotide and pA (Fig.<br />
2C). These analyses are consistent with the<br />
presence of the proposed 2,5-phosphodiester<br />
bond between C230 and U232. The sequence<br />
of the branch was verified by thin-layer<br />
chromatography (TLC) analysis of the nucleotides<br />
liberated by snake venom phosphodiesterase<br />
cleavage of purified branch nucleotide<br />
(Fig. 2D).<br />
Formation of the branched nucleotide implies<br />
a reaction mechanism in which the<br />
2OH of U232 makes a nucleophilic attack<br />
at the phosphodiester bond at IPS (Fig. 3A).<br />
To test this mechanism, we made cleavage<br />
analyses combining a ribozyme truncated in L9<br />
(157.-7) and site-specifically deoxy-substituted<br />
substrates that complemented the truncated<br />
ribozyme (7.22). Only the dU232 substrate did<br />
not support cleavage (Fig. 3B). Weak cleavage<br />
with the dA231 substrate is ascribed to a<br />
critical structural role of this nucleotide. The<br />
cleavage in the all-RNA, dC230, dA231, and<br />
dC233 substrates was by transesterification as<br />
shown by primer extension analysis (fig. S5).<br />
We have shown here that GIR1 cleaves by<br />
transesterification, not by hydrolysis as proposed<br />
previously (5). The reaction leaves a 5 fragment<br />
containing a fully active ribozyme with a<br />
3OH, and a 3 fragment in which the first and<br />
the third nucleotides are linked by a 2,5-<br />
phosphodiester bond. A 4-nt lariat was found<br />
by nuclear magnetic resonance (NMR) imaging<br />
to have an unusual structure with the sugars in<br />
the lariat ring locked in a rigid South-type<br />
conformation (14). We refer to the similarly<br />
sized lariat in Didymium as the lariat cap<br />
because it is found to cap the cellular I-Dir I<br />
mRNA (Fig. 3C). Other studies have shown<br />
that IPS1-cleaved RNA cannot be reactivated<br />
for modification at IPS2, which suggests a<br />
mechanistic coupling of the two reactions.<br />
Hydrolytic IPS1 cleavage thus appears as a<br />
failure to link the 5-phosphate of C230 to the<br />
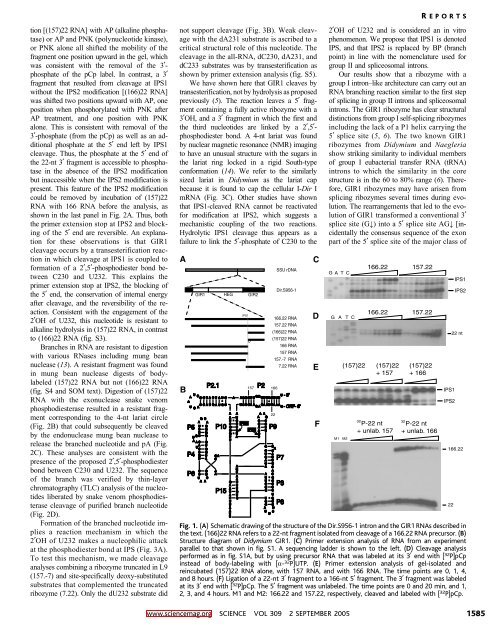
A<br />
B<br />
U<br />
A<br />
P4<br />
P6<br />
GIR1<br />
G<br />
A<br />
U<br />
C<br />
C<br />
G C<br />
A C<br />
A<br />
U<br />
C<br />
C<br />
G<br />
U<br />
C<br />
C<br />
U<br />
A<br />
C<br />
A<br />
G<br />
G<br />
G<br />
G<br />
G<br />
U<br />
G<br />
G<br />
C<br />
A<br />
G<br />
G<br />
C<br />
A<br />
P2.1<br />
G<br />
C<br />
A<br />
A<br />
A<br />
P15<br />
HEG<br />
A<br />
A<br />
U<br />
C<br />
G<br />
G<br />
G<br />
U<br />
U<br />
G<br />
A<br />
A<br />
C<br />
A<br />
C<br />
U<br />
U<br />
A<br />
A<br />
U<br />
U<br />
A<br />
U<br />
G<br />
G<br />
C<br />
C<br />
U<br />
A<br />
A<br />
G<br />
U<br />
A<br />
A<br />
C<br />
U<br />
U<br />
G<br />
U<br />
G<br />
IPS1<br />
P5 . IPS2<br />
P10<br />
.<br />
.<br />
GIR2<br />
U<br />
P2<br />
157 166<br />
A<br />
A<br />
G<br />
G<br />
A<br />
IPS1 A<br />
G<br />
C<br />
A<br />
G<br />
C<br />
C<br />
U<br />
U<br />
G<br />
C<br />
U<br />
G<br />
A<br />
C<br />
A<br />
A<br />
U<br />
U<br />
G<br />
G<br />
G<br />
U<br />
G<br />
G<br />
A<br />
U<br />
.<br />
.<br />
U<br />
SSU rDNA<br />
Dir.S956-1<br />
166.22 RNA<br />
157.22 RNA<br />
(166)22 RNA<br />
(157)22 RNA<br />
166 RNA<br />
157 RNA<br />
157.-7 RNA<br />
7.22 RNA<br />
U<br />
U<br />
A G GGU<br />
U<br />
GG<br />
- 5’<br />
A C<br />
G G U A C U A U G A<br />
GU UGGUU<br />
U<br />
A . .<br />
C C A U G A U G C U C C CA<br />
A CA AUC<br />
A<br />
G<br />
A A<br />
U<br />
A<br />
U<br />
C A<br />
- ORF- 3’<br />
22<br />
P9<br />
C<br />
A<br />
G<br />
A<br />
C<br />
U<br />
G<br />
C<br />
A<br />
C<br />
G<br />
G<br />
C<br />
C<br />
C<br />
U<br />
G<br />
C<br />
C<br />
U<br />
C<br />
P7<br />
P3<br />
P8<br />
C<br />
D<br />
E<br />
F<br />
2OH of U232 and is considered an in vitro<br />
phenomenon. We propose that IPS1 is denoted<br />
IPS, and that IPS2 is replaced by BP (branch<br />
point) in line with the nomenclature used for<br />
group II and spliceosomal introns.<br />
Our results show that a ribozyme with a<br />
group I intron–like architecture can carry out an<br />
RNA branching reaction similar to the first step<br />
of splicing in group II introns and spliceosomal<br />
introns. The GIR1 ribozyme has clear structural<br />
distinctions from group I self-splicing ribozymes<br />
including the lack of a P1 helix carrying the<br />
5 splice site (5, 6). The two known GIR1<br />
ribozymes from Didymium and Naegleria<br />
show striking similarity to individual members<br />
of group I eubacterial transfer RNA (tRNA)<br />
introns to which the similarity in the core<br />
structure is in the 60 to 80% range (6). Therefore,<br />
GIR1 ribozymes may have arisen from<br />
splicing ribozymes several times during evolution.<br />
The rearrangements that led to the evolution<br />
of GIR1 transformed a conventional 3<br />
splice site (G,) into a 5 splice site AG, [incidentally<br />
the consensus sequence of the exon<br />
part of the 5 splice site of the major class of<br />
G A T C<br />
G A T C<br />
166.22 157.22<br />
166.22<br />
157.22<br />
(157)22 (157)22 (157)22<br />
+ 157 + 166<br />
M1 M2<br />
32<br />
P-22 nt<br />
+ unlab. 157<br />
32<br />
P-22 nt<br />
+ unlab. 166<br />
IPS1<br />
IPS2<br />
22<br />
IPS1<br />
IPS2<br />
22 nt<br />
166.22<br />
Fig. 1. (A) Schematic drawing of the structure of the Dir.S956-1 intron and the GIR1 RNAs described in<br />
the text. (166)22 RNA refers to a 22-nt fragment isolated from cleavage of a 166.22 RNA precursor. (B)<br />
Structure diagram of Didymium GIR1. (C) Primer extension analysis of RNA from an experiment<br />
parallel to that shown in fig. S1. A sequencing ladder is shown to the left. (D) Cleavage analysis<br />
performed as in fig. S1A, but by using precursor RNA that was labeled at its 3 end with [ 32 P]pCp<br />
instead of body-labeling with [a- 32 P]UTP. (E) Primer extension analysis of gel-isolated and<br />
reincubated (157)22 RNA alone, with 157 RNA, and with 166 RNA. The time points are 0, 1, 4,<br />
and 8 hours. (F) Ligationofa22-nt3 fragment to a 166-nt 5 fragment. The 3 fragment was labeled<br />
at its 3 end with [ 32 P]pCp. The 5 fragment was unlabeled. The time points are 0 and 20 min, and 1,<br />
2, 3, and 4 hours. M1 and M2: 166.22 and 157.22, respectively, cleaved and labeled with [ 32 P]pCp.<br />
www.sciencemag.org SCIENCE VOL 309 2 SEPTEMBER 2005 1585