Chapter 5 Genetic Analysis of Apomixis - cimmyt
Chapter 5 Genetic Analysis of Apomixis - cimmyt
Chapter 5 Genetic Analysis of Apomixis - cimmyt
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
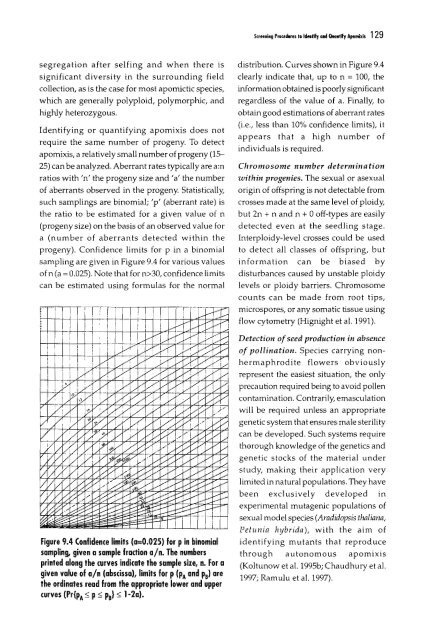
S"...iog Pro...,.. '0 Idnllfy ood a.aotlfy <strong>Apomixis</strong> 129segregation after selfing and when there issignificant diversity in the surrounding fieldcollection, as is the case for most apomictic species,which are generally polyploid, polymorphic, andhighly heterozygous.Identifying or quantifying apomixis does notrequire the same number <strong>of</strong> progeny. To detectapomixis, a relatively small number <strong>of</strong>progeny (1525) can be analyzed. Aberrant rates typically are a:nratios with 'n' the progeny size and 'a' the number<strong>of</strong> aberrants observed in the progeny. Statistically,such samplings are binomial; 'p' (aberrant rate) isthe ratio to be estimated for a given value <strong>of</strong> n(progeny size) on the basis <strong>of</strong> an observed value fora (number <strong>of</strong> aberrants detected within theprogeny). Confidence limits for p in a binomialsampling are given in Figure 9.4 for various values<strong>of</strong> n (a = 0.025). Note that for n>30, confidence limitscan be estimated using formulas for the normalFigure 9.4 (onfidence limits (a=0.025) for pin binomialsampling, given a sample fraction a/n. The numbersprinted along the curves indicate the sample size, n. For agiven value <strong>of</strong> aln (abscissa), limits for p (PA and PB) arethe ordinates read from the appropriate lower and uppercurves (Pr{PA ~ p ~ PB} ~ 1-2a).distribution. Curves shown in Figure 9.4clearly indicate that, up to n = 100, theinformation obtained is poorly signjficantregardless <strong>of</strong> the value <strong>of</strong> a. Finally, toobtain good estimations <strong>of</strong> aberrant rates(i.e., less than 10% confidence limits), itappears that a high number <strong>of</strong>individuals is required.Chromosome number determinationwithin progenies. The sexual or ;:lsexualorigin <strong>of</strong> <strong>of</strong>fspring is not detectable fromcrosses made at the same level <strong>of</strong> ploidy,but 2n + nand n + 0 <strong>of</strong>f-types are easilydetected even at the seedling stage.lnterploidy-Ievel crosses could be usedto detect all classes <strong>of</strong> <strong>of</strong>fspring, butinformation can be biased bydisturbances caused by unstable ploidylevels or ploidy barriers. Chromosomecounts can be made from root tips,microspores, or any somatic tissue usingflow cytometry (Hignight et al. 1991).Detection <strong>of</strong> seed production in absence<strong>of</strong> pollination. Species carrying nonhermaphroditeflowers obviouslyrepresent the easiest situation, the onlyprecaution required being to avoid pollencontamination. Contrarily, emasculationwill be required unless an appropriategenetic system that ensures male sterilitycan be developed. Such systems requirethorough knowledge <strong>of</strong> the genetics andgenetic stocks <strong>of</strong> the material understudy, making their application verylimited in natural populations. They havebeen exclusively developed Inexperimental mutagenic populations <strong>of</strong>sexual model species (Aradidopsis tha/ulna,Petllnia hybrida), with the aim <strong>of</strong>identifying mutants that reproducethrough autonomous apomixis(Koltunow et al. 1995b; Chaudhury et al.1997; Ramulu et al. 1997).