Chapter 5 Genetic Analysis of Apomixis - cimmyt
Chapter 5 Genetic Analysis of Apomixis - cimmyt
Chapter 5 Genetic Analysis of Apomixis - cimmyt
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
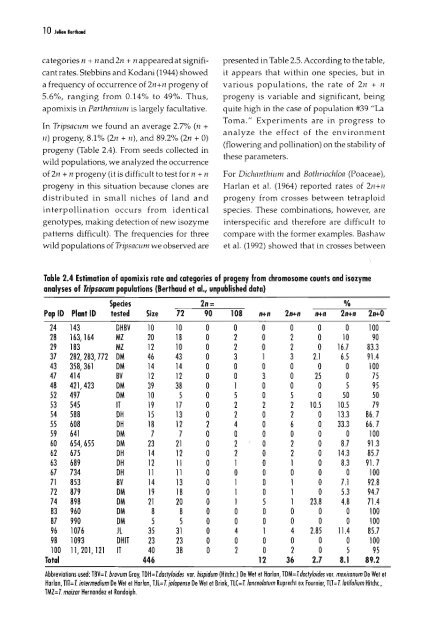
10M.. Be,th.dcategories n + nand 2n + n appeared at signifi- presented in Table 2.5. According to the table,cant rates. Stebbins and Kodani (1944) showed it appears that within one species, but ina frequency <strong>of</strong> occurrence <strong>of</strong> 2n+n progeny <strong>of</strong> various populations, the rate <strong>of</strong> 2n + n5.6%, ranging from 0.14% to 49%. Thus, progeny is variable and significant, beingapomixis in Parthenillm is largely facultative. quite high in the case <strong>of</strong> population #39 "LaToma." Experiments are in progress toIn Tripsawm we found an average 2.7% (n +analyze the effect <strong>of</strong> the environment11) progeny, 8.1% (2/1 + 11), and 89.2% (2n + 0)(flowering and pollination) on the stability <strong>of</strong>progeny (Table 2.4). From seeds collected inthese parameters.wild populations, we analyzed the occurrence<strong>of</strong> 2n + n progeny (it is difficult to test for n + n For Dichanthium and Bothriochloa (Poaceae),progeny in this situation because clones are Harlan et al. (1964) reported rates <strong>of</strong> 211+11distributed in small niches <strong>of</strong> land and progeny from crosses between tetraploidinterpollination occurs from identical species. These combinations, however, aregenotypes, making detection <strong>of</strong> new isozyme interspecific and therefore are difficult topatterns difficult). The frequencies for three compare with the former examples. Bashawwild populations <strong>of</strong> TripsaCllm we observed are et al. (1992) showed that in crosses betweenTable 2.4 Estimation <strong>of</strong> apomixis rate and categories <strong>of</strong> progeny from chromosome counts and isozymeanalyses <strong>of</strong> Tripsacum populations (Berthoud et 01., unpublished data)Species 2n= %Pop ID Plant ID tested Size 72 90 108 n+n 2n+n n+n 2n+n 2n+024 143 DHBV 10 10 0 0 0 a a a 10028 163,164 MZ 20 18 a 2 a 2 a 10 9029 183 MZ 12 10 a 2 a 2 a 16.7 83.337 282, 283, 772 OM 46 43 a 3 1 3 2.1 6.5 91.443 358,361 OM 14 14 a a a a a a 10047 414 BV 12 12 a a 3 a 25 a 7548 421,423 OM 39 38 a 1 a a a 5 9552 497 OM 10 5 a 5 a 5 a 50 5053 545 II 19 17 a 2 2 2 10.5 10.5 7954 588 OH 15 13 a 2 a 2 a 13.3 86. 755 608 OH 18 12 2 4 a 6 a 33.3 66. 759 641 OM 7 7 a a a a a a 10060 654, 655 OM 23 21 a 2 a 2 a 8.7 91.362 675 OH 14 12 a 2 a 2 a 14.3 85.763 689 OH 12 11 a 1 a 1 a 8.3 91.767 734 OH 11 11 a a a a a a 10071 853 BV 14 13 a 1 a 1 a 7.1 92.872 879 OM 19 18 a 1 a 1 a 5.3 94.774 898 OM 21 20 a 1 5 1 23.8 4.8 71.4B3 960 OM 8 8 a a a a a a 10087 990 OM 5 5 a a a a a a lOa96 1076 Jl 35 31 a 4 1 4 2.85 11.4 85.798 1093 OHII 23 23 a a a a a a 100100 11,201,121 IT 40 38 a 2 a 2 a 5 95Total 446 12 36 2.7 8.1 89.2Abbreviations used: TBV=T. bravum Gray, TDH=T.dactylaides var. hispidum (Hitchc.) De Wet et Harlan, TDM=T.dactylaides var. mex;canum De WeI etHarlan, TIT=T. in/ermedium De Wet et Harlan, IJl=T. jalapense De Wet et Brink, TlC=T.lancealalum Ruprecht ex Fournier, TlT=T.lalifalium Hilchc.,TMZ= T. maizar Hernandez el Randolph.