primary prevention of coeliac disease - Associazione Italiana ...
primary prevention of coeliac disease - Associazione Italiana ...
primary prevention of coeliac disease - Associazione Italiana ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
COELIAC DISEASE IN THE SAHARAWI<br />
35<br />
the border between the European and the African range <strong>of</strong> variability. Thus, the genetic<br />
structure <strong>of</strong> this population is consistent with its geographical location.<br />
The association <strong>of</strong> the DQ molecules with CD in the Saharawi population<br />
We first established the sibling lifetime risk/population prevalence ratio, ls in our<br />
Saharawi sample set, that was equal to 3 (16.7%/5.6%). Because <strong>of</strong> the high <strong>disease</strong><br />
prevalence, the degree <strong>of</strong> familial clustering was lower in the Saharawi than in<br />
Caucasian populations ls = ~30).<br />
Next, we established the association <strong>of</strong> the haplotypes and genotypes at the main<br />
<strong>disease</strong> superlocus HLA-DQB1-DQA1 in this population. The distribution <strong>of</strong> the<br />
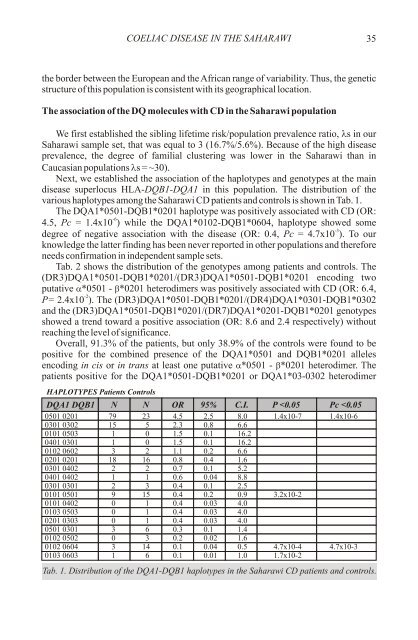
various haplotypes among the Saharawi CD patients and controls is shown in Tab. 1.<br />
The DQA1*0501-DQB1*0201 haplotype was positively associated with CD (OR:<br />
-6<br />
4.5, Pc = 1.4x10 ) while the DQA1*0102-DQB1*0604, haplotype showed some<br />
-3<br />
degree <strong>of</strong> negative association with the <strong>disease</strong> (OR: 0.4, Pc = 4.7x10 ). To our<br />
knowledge the latter finding has been never reported in other populations and therefore<br />
needs confirmation in independent sample sets.<br />
Tab. 2 shows the distribution <strong>of</strong> the genotypes among patients and controls. The<br />
(DR3)DQA1*0501-DQB1*0201/(DR3)DQA1*0501-DQB1*0201 encoding two<br />
putative a*0501 - b*0201 heterodimers was positively associated with CD (OR: 6.4,<br />
-2<br />
P= 2.4x10 ). The (DR3)DQA1*0501-DQB1*0201/(DR4)DQA1*0301-DQB1*0302<br />
and the (DR3)DQA1*0501-DQB1*0201/(DR7)DQA1*0201-DQB1*0201 genotypes<br />
showed a trend toward a positive association (OR: 8.6 and 2.4 respectively) without<br />
reaching the level <strong>of</strong> significance.<br />
Overall, 91.3% <strong>of</strong> the patients, but only 38.9% <strong>of</strong> the controls were found to be<br />
positive for the combined presence <strong>of</strong> the DQA1*0501 and DQB1*0201 alleles<br />
encoding in cis or in trans at least one putative a*0501 - b*0201 heterodimer. The<br />
patients positive for the DQA1*0501-DQB1*0201 or DQA1*03-0302 heterodimer<br />
HAPLOTYPES Patients Controls<br />
DQA1 DQB1 N N OR 95% C.I. P