4. DISCUSSIONMolecular techniques are used to study symbionts and parasites, includingplant pathog<strong>en</strong>s and lich<strong>en</strong>ized and mycorrhizal fungi. However,Laboulb<strong>en</strong>iales show particular difficulties wh<strong>en</strong> it comes to DNA extraction:their microscopic size, the finite number of thick-walled cells in the thallus andfrequ<strong>en</strong>tly heavily melanized regions, thus difficult to break op<strong>en</strong>. Therefore,attempts to amplify ITS sequ<strong>en</strong>ces of Laboulb<strong>en</strong>iales failed. Manycontaminations were found instead.For all 48 specim<strong>en</strong>s, ITS amplification following the differ<strong>en</strong>t protocols (WEIR &BLACKWELL, 2001a; WEIR & BLACKWELL, 2001b; 2.2. MORPHOLOGICAL PROTOCOL ANDDNA EXTRACTION) was not successful in releasing laboulb<strong>en</strong>ialean DNA. For 12specim<strong>en</strong>s, amplification with ITS5/ITS4-A led to sequ<strong>en</strong>ces that could be usedfor comparison with G<strong>en</strong>Bank. Matches were obtained from differ<strong>en</strong>tascomycotan classes: Dothideomycetes, Pezizomycetes and Saccharomycetes.For one specim<strong>en</strong> [flag1146], there ev<strong>en</strong> was a 99% similarity matchwith Dioscorea polystachya, a member of the Archaeplastida (while fungi areOpisthokonta). One specim<strong>en</strong> amplified using ITS5/ITS2 [flag1151] matched inG<strong>en</strong>Bank with Myc<strong>en</strong>a amabilissima (Basidiomycota, Agaricomycetes). Thephylog<strong>en</strong>y of the most common resulting matches in G<strong>en</strong>Bank is pres<strong>en</strong>ted inFigure X below.SpermatophytaAscomycota, DothideomycetesAscomycota, PezizomycetesBasidiomycota, AgaricomycetesAscomycota, SaccharomycetesFigure X: Phylog<strong>en</strong>y of the most common resulting matches in G<strong>en</strong>Bank, based on 18S rDNA. Thephylog<strong>en</strong>etic tree is the result of a neighbor joining analysis of a dataset with 15 sequ<strong>en</strong>ces.Sequ<strong>en</strong>ces (with accession number betwe<strong>en</strong> brackets) are (from top to bottom) Dioscoreapolystachya (FJ860063), Cladosporium sp. (GU214631), Davidiella tassiana (EU343349),Cladosporium cladoporioides (AY251074), Cladosporium ossifragi (EF679382), Davidiella macrospora(EU167591), Choiromyces v<strong>en</strong>osus (AF435827), Myc<strong>en</strong>a amabilissima (DQ490644), Pichia sp.(HM627157), Candida ferm<strong>en</strong>ticar<strong>en</strong>s (FM178353), Pichia castillae (DQ409168), Pichia media(DQ409170), Pichia stipitis (CP000497), Debaromyces hans<strong>en</strong>ii (GQ458025) and Debaromycespseudopolymorphus (EF198011).P a g e | 41
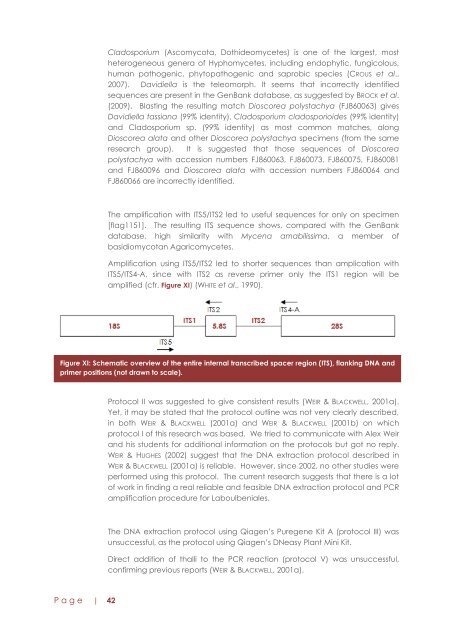
Cladosporium (Ascomycota, Dothideomycetes) is one of the largest, mostheterog<strong>en</strong>eous g<strong>en</strong>era of Hyphomycetes, including <strong>en</strong>dophytic, fungicolous,human pathog<strong>en</strong>ic, phytopathog<strong>en</strong>ic and saprobic species (CROUS et al.,2007). Davidiella is the teleomorph. It seems that incorrectly id<strong>en</strong>tifiedsequ<strong>en</strong>ces are pres<strong>en</strong>t in the G<strong>en</strong>Bank database, as suggested by BROCK et al.(2009). Blasting the resulting match Dioscorea polystachya (FJ860063) givesDavidiella tassiana (99% id<strong>en</strong>tity), Cladosporium cladosporioides (99% id<strong>en</strong>tity)and Cladosporium sp. (99% id<strong>en</strong>tity) as most common matches, alongDioscorea alata and other Dioscorea polystachya specim<strong>en</strong>s (from the sameresearch group). It is suggested that those sequ<strong>en</strong>ces of Dioscoreapolystachya with accession numbers FJ860063, FJ860073, FJ860075, FJ860081and FJ860096 and Dioscorea alata with accession numbers FJ860064 andFJ860066 are incorrectly id<strong>en</strong>tified.The amplification with ITS5/ITS2 led to useful sequ<strong>en</strong>ces for only on specim<strong>en</strong>[flag1151]. The resulting ITS sequ<strong>en</strong>ce shows, compared with the G<strong>en</strong>Bankdatabase, high similarity with Myc<strong>en</strong>a amabilissima, a member ofbasidiomycotan Agaricomycetes.Amplification using ITS5/ITS2 led to shorter sequ<strong>en</strong>ces than amplication withITS5/ITS4-A, since with ITS2 as reverse primer only the ITS1 region will beamplified (cfr. Figure XI) (WHITE et al., 1990).Figure XI: Schematic overview of the <strong>en</strong>tire internal transcribed spacer region (ITS), flanking DNA andprimer positions (not drawn to scale).Protocol II was suggested to give consist<strong>en</strong>t results (WEIR & BLACKWELL, 2001a).Yet, it may be stated that the protocol outline was not very clearly described,in both WEIR & BLACKWELL (2001a) and WEIR & BLACKWELL (2001b) on whichprotocol I of this research was based. We tried to communicate with Alex Weirand his stud<strong>en</strong>ts for additional information on the protocols but got no reply.WEIR & HUGHES (2002) suggest that the DNA extraction protocol described inWEIR & BLACKWELL (2001a) is reliable. However, since 2002, no other studies wereperformed using this protocol. The curr<strong>en</strong>t research suggests that there is a lotof work in finding a real reliable and feasible DNA extraction protocol and PCRamplification procedure for Laboulb<strong>en</strong>iales.The DNA extraction protocol using Qiag<strong>en</strong>‟s Pureg<strong>en</strong>e Kit A (protocol III) wasunsuccessful, as the protocol using Qiag<strong>en</strong>‟s DNeasy Plant Mini Kit.Direct addition of thalli to the PCR reaction (protocol V) was unsuccessful,confirming previous reports (WEIR & BLACKWELL, 2001a).P a g e | 42
- Page 3: LABOULBENIALESEXPLORING AND TESTING
- Page 7 and 8: PART IVPRELIMINARY CHECKLIST OF LAB
- Page 9 and 10: SAMENVATTINGINLEIDINGLaboulbeniales
- Page 11 and 12: PART IGENERAL INTRODUCTIONTHESIS OU
- Page 13 and 14: fifth volume, therefore the sixth v
- Page 16 and 17: 1.3.3. THE PERITHECI UMAscospores o
- Page 18 and 19: The identity of appendages, togethe
- Page 20 and 21: Figure IV: Position of Laboulbeniom
- Page 22 and 23: LUMBSCH & HUHNDORF (2007) distingui
- Page 25 and 26: Substrate is the intermediate facto
- Page 27: CAVARA (1899, ref. in BENJAMIN, 197
- Page 35 and 36: 1. INTRODUCTION1.1. DIFFICULTIES FO
- Page 37 and 38: 2. MATERIALS & METHODS2.1. FUNGUS,
- Page 39 and 40: 2.2.6. PROTOCOL V: DIRECT PCROne th
- Page 41 and 42: P a g e | 38chance. Thus, the lower
- Page 43: coll1133 Laboulbenia collae IV 608l
- Page 47 and 48: 5. CONCLUSION AND SUGGESTIONSMany a
- Page 49 and 50: 1. INTRODUCTION1.1. FORENSIC ENTOMO
- Page 51 and 52: Different groups of carrion beetles
- Page 53 and 54: The 1.319 ha Meerdaal Forest has be
- Page 55 and 56: 2.2. MEERDAAL FOREST(DA TA NA TI O
- Page 57 and 58: approximately 2.700 are found in Eu
- Page 59 and 60: Order ColeopteraSuborder PolyphagaI
- Page 61 and 62: Dermestidae 8 2 23 (3) / 22 (4)Elat
- Page 63 and 64: Table XXIV: Family Cerambycidae fro
- Page 65 and 66: Table XXVIII: Family Dermestidae fr
- Page 67 and 68: Adults prey on other flower-visitin
- Page 69 and 70: Table XXXV: Family Scarabaeidae fro
- Page 71 and 72: Creophilus Samouellemaxillosus (Lin
- Page 73 and 74: 4.3. LABOULBENIALES OF CARRION BEET
- Page 75 and 76: The knowledge about the preferences
- Page 77 and 78: Table XLII: Comparison of carrion b
- Page 79 and 80: 5. CONCLUSION AND SUGGESTIONSNo spe
- Page 81 and 82: 1. INTRODUCTION1.1. NATURAL LANDSCA
- Page 83 and 84: In the 40s, Middelhoek studied and
- Page 85 and 86: 2. MATERIALS & METHODS2.1. HOSTS2.1
- Page 87 and 88: Screening of the insects was done w
- Page 89 and 90: 3. RESULTS3.1. PARASITE-HOST LISTA
- Page 91 and 92: Figure XV: Detail of thallus of Lab
- Page 93 and 94: isodiametric or slightly elongate,
- Page 95 and 96:
occurs in the lower receptacle (cel
- Page 97 and 98:
Discussion:Two species of the genus
- Page 99 and 100:
anches (15-)34-38(-40) µm long, te
- Page 101 and 102:
5. CONCLUSION AND SUGGESTIONSIn The
- Page 103 and 104:
P a g e | 100ADRIAENS, T. & GYSELS,
- Page 105 and 106:
transmission, habitat preference an
- Page 107 and 108:
STALPERS, J.A., VILGALYS, R., AIME,
- Page 109 and 110:
MELIS, C., TEURLINGS, I. LINNELL, J
- Page 111 and 112:
SCHILTHUIZEN, M. & VALLENDUUK, H. (