computer modeling in molecular biology.pdf
computer modeling in molecular biology.pdf
computer modeling in molecular biology.pdf
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
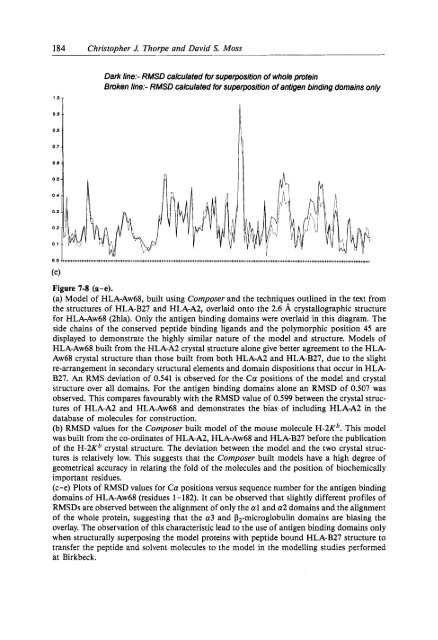
184 Christopher J. Thorpe and David S. Moss'.aTDark l<strong>in</strong>e:- RMSD calculated for superposition of whole prote<strong>in</strong>Broken l<strong>in</strong>e:- RMSD calculated for superposition of antigen b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong>s only0.00.80.7 -0.6 -0.0 0(elFigure 7-8 (a-e).(a) Model of HLA-Aw68, built us<strong>in</strong>g Composer and the techniques outl<strong>in</strong>ed <strong>in</strong> the text fromthe structures of HLA-B27 and HLA-A2, overlaid onto the 2.6 A crystallographic structurefor HLA-Aw68 (2hla). Only the antigen b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong>s were overlaid <strong>in</strong> this diagram. Theside cha<strong>in</strong>s of the conserved peptide b<strong>in</strong>d<strong>in</strong>g ligands and the polymorphic position 45 aredisplayed to demonstrate the highly similar nature of the model and structure. Models ofHLA-Aw68 built from the HLA-A2 crystal structure alone give better agreement to the HLA-Aw68 crystal structure than those built from both HLA-A2 and HLA-B27, due to the slightre-arrangement <strong>in</strong> secondary structural elements and doma<strong>in</strong> dispositions that occur <strong>in</strong> HLA-B27. An RMS deviation of 0.541 is observed for the Ca positions of the model and crystalstructure over all doma<strong>in</strong>s. For the antigen b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong>s alone an RMSD of 0.507 wasobserved. This compares favourably with the RMSD value of 0.599 between the crystal structuresof HLA-A2 and HLA-Aw68 and demonstrates the bias of <strong>in</strong>clud<strong>in</strong>g HLA-A2 <strong>in</strong> thedatabase of molecules for construction.(b) RMSD values for the Composer built model of the mouse molecule H-2Kb. This modelwas built from the co-ord<strong>in</strong>ates of HLA-A2, HLA-Aw68 and HLA-B27 before the publicationof the H-2Kb crystal structure. The deviation between the model and the two crystal structuresis relatively low. This suggests that the Composer built models have a high degree ofgeometrical accuracy <strong>in</strong> relat<strong>in</strong>g the fold of the molecules and the position of biochemicallyimportant residues.(c-e) Plots of RMSD values for Ca positions versus sequence number for the antigen b<strong>in</strong>d<strong>in</strong>gdoma<strong>in</strong>s of HLA-Aw68 (residues 1 - 182). It can be observed that slightly different profiles ofRMSDs are observed between the alignment of only the a1 and a2 doma<strong>in</strong>s and the alignmentof the whole prote<strong>in</strong>, suggest<strong>in</strong>g that the a3 and Pz-microglobul<strong>in</strong> doma<strong>in</strong>s are bias<strong>in</strong>g theoverlay. The observation of this characteristic lead to the use of antigen b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong>s onlywhen structurally superpos<strong>in</strong>g the model prote<strong>in</strong>s with peptide bound HLA-B27 structure totransfer the peptide and solvent molecules to the model <strong>in</strong> the modell<strong>in</strong>g studies performedat Birkbeck.