computer modeling in molecular biology.pdf
computer modeling in molecular biology.pdf
computer modeling in molecular biology.pdf
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
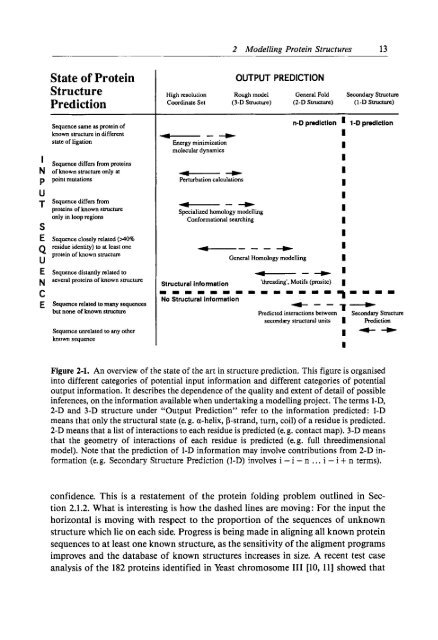
2 Modell<strong>in</strong>g Prote<strong>in</strong> Structures 13State of Prote<strong>in</strong>StructurePredictionSequence same as prote<strong>in</strong> ofknown sbucture <strong>in</strong> differentstate of ligation’ Sequence differs from prote<strong>in</strong>sN of known smcture only atp p<strong>in</strong>t mutationsUT Sequence differs Fromprote<strong>in</strong>s of known structureonly <strong>in</strong> loop regionsSE Sequence closely related ( ~ 0 %residue identity) to at least oneQ prote<strong>in</strong> of known structureUE Sequence distantly relsled toseveral prote<strong>in</strong>s of known structureNCE Squence related to many sequencesbut none of known structureSequence unrelated to any otherknown sequenceOUTPUT PREDICTIONHigh resolution Rough model General Fold Scoondary StructureCoord<strong>in</strong>ate Set (3-D Structure) (2-D Structure) (I-D Structure)--+Energy m<strong>in</strong>imization<strong>molecular</strong> dynamicsn-D prediction I 1-D predictiona--+Specialized homology modell<strong>in</strong>gConformational search<strong>in</strong>g----+ IGeneral Homology modell<strong>in</strong>g I--+ ’Structural <strong>in</strong>formation ‘thread<strong>in</strong>g’. Motifs (pmsite) Im m 1 1 1 m m m 1 1 m 1 1 =No Structural Information7====+--T---Predicted <strong>in</strong>teractions between Secondary Structuresecondary structural units I PredictionIIFigure 2-1. An overview of the state of the art <strong>in</strong> structure prediction. This figure is organised<strong>in</strong>to different categories of potential <strong>in</strong>put <strong>in</strong>formation and different categories of potentialoutput <strong>in</strong>formation. It describes the dependence of the quality and extent of detail of possible<strong>in</strong>ferences, on the <strong>in</strong>formation available when undertak<strong>in</strong>g a modell<strong>in</strong>g project. The terms 1-42-D and 3-D structure under “Output Prediction” refer to the <strong>in</strong>formation predicted: 1-Dmeans that only the structural state (e. g. a-helix, P-strand, turn, coil) of a residue is predicted.2-D means that a list of <strong>in</strong>teractions to each residue is predicted (e. g. contact map). 3-D meansthat the geometry of <strong>in</strong>teractions of each residue is predicted (e. g. full threedimensionalmodel). Note that the prediction of 1-D <strong>in</strong>formation may <strong>in</strong>volve contributions from 2-D <strong>in</strong>formation(e. g. Secondary Structure Prediction (I-D) <strong>in</strong>volves i - i - n . . . i - i + n terms).confidence. This is a restatement of the prote<strong>in</strong> fold<strong>in</strong>g problem outl<strong>in</strong>ed <strong>in</strong> Section2.1.2. What is <strong>in</strong>terest<strong>in</strong>g is how the dashed l<strong>in</strong>es are mov<strong>in</strong>g: For the <strong>in</strong>put thehorizontal is mov<strong>in</strong>g with respect to the proportion of the sequences of unknownstructure which lie on each side. Progress is be<strong>in</strong>g made <strong>in</strong> align<strong>in</strong>g all known prote<strong>in</strong>sequences to at least one known structure, as the sensitivity of the aligment programsimproves and the database of known structures <strong>in</strong>creases <strong>in</strong> size. A recent test caseanalysis of the 182 prote<strong>in</strong>s identified <strong>in</strong> Yeast chromosome I11 [lo, 111 showed that