Annual Scientific Report 2015
EMBL_EBI_ASR_2015_DigitalEdition
EMBL_EBI_ASR_2015_DigitalEdition
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Claire O’Donovan<br />
Protein Function Content<br />
BSc (Hons) in Biochemistry, University College<br />
Cork, 1992. Diploma in Computer Science<br />
University College Cork, 1993.<br />
Future plans<br />
In 2016 we will continue work on a ‘gold-standard’<br />
dataset across the taxonomic range, with a particular<br />
focus on the UniProt proteomes set to fully address<br />
the requirements of the biochemical community. We<br />
will also continue to expand and refine our Ensembl<br />
and Genome Reference Consortium collaborations to<br />
ensure that UniProtKB provides the most appropriate<br />
gene-centric view of the protein space, allowing a<br />
cleaner and more logical mapping of gene and genomic<br />
resources to UniProtKB. We will continue to co-operate<br />
with diverse data providers (e.g., Ensembl, RefSeq,<br />
PRIDE) to integrate relevant genome and proteome<br />
information, and will import variation information from<br />
COSMIC. We also plan to extend our nomenclature<br />
collaborations to include higher-level organisms.<br />
We will prioritise the extraction of experimental data<br />
from the literature and extend our use of data-mining<br />
methods to identify scientific literature of particular<br />
interest with regard to our annotation priorities. We<br />
are committed to expanding UniRule by extending<br />
the number and range of rules with additional curator<br />
resources, both internal and external, and providing<br />
these rules to external collaborators for use in their<br />
systems.<br />
In 2016 we also plan to extend the scope of GO<br />
annotation to encompass entities other than proteins, in<br />
particular RNA and protein complexes.<br />
At EMBL since 1993, at EMBL-EBI since 1994.<br />
Team Leader since 2009.<br />
Selected publications<br />
Alam-Faruque Y, Hill DP, Dimmer EC, et al. (2014)<br />
Representing kidney development using the gene<br />
ontology. PLoS One 9:e99864<br />
Alpi E, Griss J, da Silva AW, et al. (2014) Analysis of the<br />
tryptic search space in UniProt databases. Proteomics<br />
15:48-57<br />
Huntley RP, Sawford T, Martin MJ and O’Donovan C<br />
(2014) Understanding how and why the Gene Ontology<br />
and its annotations evolve: the GO within UniProt.<br />
Gigascience 3:4<br />
Huntley RP, Sawford T, Mutowo-Meullenet P, et al.<br />
(2014) The GOA database: gene ontology annotation<br />
updates for <strong>2015</strong>. Nucl Acids Res 43(database<br />
issue):d1057-63<br />
Poux S, Magrane M, Arighi CN, et al. (2014) Expert<br />
curation in UniProtKB: a case study on dealing with<br />
conflicting and erroneous data. Database (Oxford) 2014:<br />
bau016<br />
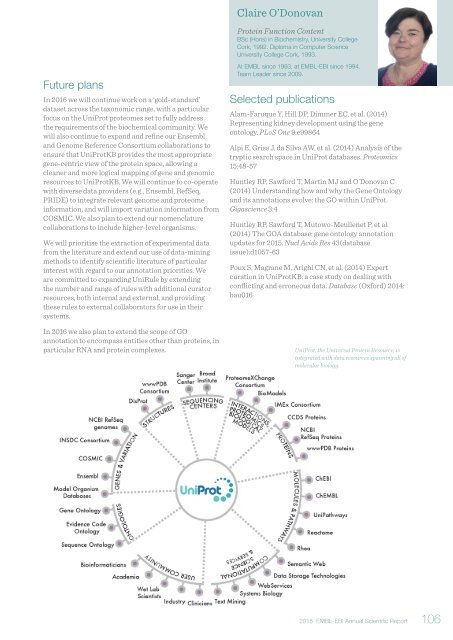
UniProt, the Universal Protein Resource, is<br />
integrated with data resources spanning all of<br />
molecular biology.<br />
<strong>2015</strong> EMBL-EBI <strong>Annual</strong> <strong>Scientific</strong> <strong>Report</strong> 106