Annual Scientific Report 2015
EMBL_EBI_ASR_2015_DigitalEdition
EMBL_EBI_ASR_2015_DigitalEdition
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Anton Enright<br />
PhD in Computational Biology, University of<br />
Cambridge, 2003. Postdoctoral research at<br />
Memorial Sloan-Kettering Cancer Center,<br />
New York.<br />
At EMBL-EBI since 2008.<br />
cortex<br />
9<br />
106<br />
55<br />
65<br />
106<br />
1<br />
5<br />
1<br />
117<br />
18<br />
3<br />
6<br />
537<br />
23<br />
2529<br />
1412 1 13<br />
brain<br />
5<br />
8<br />
2<br />
54<br />
100<br />
7 1<br />
7 1<br />
2<br />
161<br />
50 6<br />
70<br />
4<br />
2<br />
49<br />
2<br />
33<br />
41<br />
2<br />
5<br />
1<br />
3<br />
4<br />
2<br />
547<br />
435<br />
2<br />
32<br />
1546 4 1184 122<br />
29<br />
1210<br />
1019 9<br />
cerebellum<br />
20<br />
141<br />
2<br />
77<br />
99<br />
6<br />
92<br />
8 7<br />
5<br />
181 8<br />
3<br />
41<br />
8<br />
4<br />
1<br />
92<br />
180 1 18<br />
49<br />
200<br />
1<br />
13<br />
1<br />
1<br />
1<br />
16<br />
11<br />
421<br />
302<br />
1321<br />
1261<br />
2434 87<br />
49<br />
37<br />
71<br />
1825<br />
4<br />
258<br />
118<br />
24<br />
111<br />
8<br />
1<br />
101<br />
9<br />
4<br />
84<br />
2<br />
44<br />
3<br />
1<br />
1<br />
293<br />
27<br />
994956<br />
11 2<br />
heart<br />
19<br />
3<br />
19<br />
1<br />
3<br />
1<br />
1<br />
17<br />
86<br />
115<br />
1<br />
7<br />
6<br />
10<br />
1<br />
5<br />
4<br />
1<br />
20<br />
63 77<br />
1<br />
kidney<br />
1<br />
2<br />
30<br />
2<br />
10<br />
7<br />
1<br />
2<br />
5<br />
1<br />
129<br />
15<br />
247431<br />
3<br />
52<br />
31<br />
38<br />
15<br />
21<br />
41<br />
9<br />
1<br />
2<br />
3<br />
118<br />
2<br />
405<br />
637 10 16<br />
liver<br />
1<br />
11<br />
11<br />
1<br />
9<br />
12<br />
3<br />
2<br />
61<br />
3<br />
165<br />
4<br />
352<br />
12<br />
11<br />
2<br />
22<br />
5<br />
5<br />
1<br />
1<br />
64<br />
5<br />
96 152<br />
5<br />
olfactory bulb<br />
8<br />
101<br />
3<br />
5<br />
5<br />
108<br />
1<br />
43<br />
8<br />
1<br />
2<br />
7<br />
3<br />
14<br />
3<br />
4<br />
5<br />
20<br />
394<br />
1<br />
1002<br />
1055 20<br />
4<br />
testes<br />
1<br />
171<br />
29<br />
82<br />
11<br />
54<br />
14<br />
5 8 2<br />
22<br />
12<br />
1<br />
2<br />
5<br />
1<br />
57<br />
99<br />
31<br />
32 1 23<br />
10<br />
4<br />
3<br />
6<br />
1<br />
2<br />
1<br />
401<br />
86<br />
36<br />
1256<br />
324<br />
1905 37 80<br />
11 11<br />
467<br />
40<br />
21<br />
1<br />
11<br />
15<br />
14<br />
1<br />
3<br />
1<br />
83<br />
5<br />
224281<br />
5<br />
34709696 34745393 34781090 34816788<br />
Grk4<br />
Htt<br />
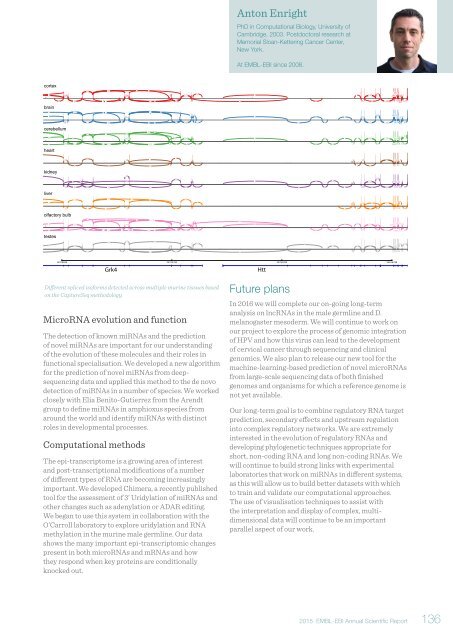
Different spliced isoforms detected across multiple murine tissues based<br />
on the CaptureSeq methodology.<br />
MicroRNA evolution and function<br />
The detection of known miRNAs and the prediction<br />
of novel miRNAs are important for our understanding<br />
of the evolution of these molecules and their roles in<br />
functional specialisation. We developed a new algorithm<br />
for the prediction of novel miRNAs from deepsequencing<br />
data and applied this method to the de novo<br />
detection of miRNAs in a number of species. We worked<br />
closely with Elia Benito-Gutierrez from the Arendt<br />
group to define miRNAs in amphioxus species from<br />
around the world and identify miRNAs with distinct<br />
roles in developmental processes.<br />
Computational methods<br />
The epi-transcriptome is a growing area of interest<br />
and post-transcriptional modifications of a number<br />
of different types of RNA are becoming increasingly<br />
important. We developed Chimera, a recently published<br />
tool for the assessment of 3’ Uridylation of miRNAs and<br />
other changes such as adenylation or ADAR editing.<br />
We began to use this system in collaboration with the<br />
O’Carroll laboratory to explore uridylation and RNA<br />
methylation in the murine male germline. Our data<br />
shows the many important epi-transcriptomic changes<br />
present in both microRNAs and mRNAs and how<br />
they respond when key proteins are conditionally<br />
knocked out.<br />
Future plans<br />
In 2016 we will complete our on-going long-term<br />
analysis on lncRNAs in the male germline and D.<br />
melanogaster mesoderm. We will continue to work on<br />
our project to explore the process of genomic integration<br />
of HPV and how this virus can lead to the development<br />
of cervical cancer through sequencing and clinical<br />
genomics. We also plan to release our new tool for the<br />
machine-learning-based prediction of novel microRNAs<br />
from large-scale sequencing data of both finished<br />
genomes and organisms for which a reference genome is<br />
not yet available.<br />
Our long-term goal is to combine regulatory RNA target<br />
prediction, secondary effects and upstream regulation<br />
into complex regulatory networks. We are extremely<br />
interested in the evolution of regulatory RNAs and<br />
developing phylogenetic techniques appropriate for<br />
short, non-coding RNA and long non-coding RNAs. We<br />
will continue to build strong links with experimental<br />
laboratories that work on miRNAs in different systems,<br />
as this will allow us to build better datasets with which<br />
to train and validate our computational approaches.<br />
The use of visualisation techniques to assist with<br />
the interpretation and display of complex, multidimensional<br />
data will continue to be an important<br />
parallel aspect of our work.<br />
<strong>2015</strong> EMBL-EBI <strong>Annual</strong> <strong>Scientific</strong> <strong>Report</strong> 136