Annual Scientific Report 2015
EMBL_EBI_ASR_2015_DigitalEdition
EMBL_EBI_ASR_2015_DigitalEdition
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Molecular Interactions<br />
The Molecular Interactions Team provides public-domain interaction data, the<br />
use of which ranges from network analysis of large-scale datasets to obtaining<br />
a detailed understanding of specific protein-binding interfaces. The team also<br />
produces the Complex Portal, a reference resource for macromolecular complexes.<br />
The Molecular Interactions Team, created in April<br />
<strong>2015</strong>, was previously part of the Proteomics Services<br />
team. We have responsibility for the long-established<br />
IntAct molecular interaction database, and for the<br />
more recent Complex Portal. Our team will continue to<br />
support the molecular interaction community standards<br />
published by the HUPO Proteomics Standards<br />
Initiative, and contribute to their further development.<br />
We actively encourage the direct deposition of data<br />
by data producers as part of the publication process,<br />
as this provides the best opportunities to ensure the<br />
information is represented accurately in the database.<br />
IntAct is the major deposition database for groups<br />
generating high-throughput, two-hybrid and affinitypurified<br />
Mass Spectrometry data. We curate small-scale<br />
data from the scientific literature, with strict qualitycontrol<br />
procedures to ensure we provide the highest<br />
quality reference datasets.<br />
The Complex Portal has increased in content<br />
and coverage since its launch in 2014. Successful<br />
collaborations with groups such as the Saccharomyces<br />
Genome Database have led to a shared curation effort<br />
and broadened community access to domain-specific<br />
expertise.<br />
Major achievements<br />
A To meet the changing demands that arise from<br />
the ever-growing complexity of interaction data, we<br />
updated the standard format for molecular interaction<br />
data (PSI-MI XML3.0). The format now allows for the<br />
description of allosteric interactions, dynamic data<br />
and protein-complex data abstracted from multiple<br />
publications. It also offers improved representation of<br />
the effect of a mutant or variant on an interaction.<br />
We are major contributors to the IMEx Consortium of<br />
interaction databases, which manages curation efforts<br />
over multiple resources. As of December <strong>2015</strong>, IntAct<br />
database made almost 600,000 binary interaction<br />
evidences publicly available. Its database<br />
infrastructure is shared as a curation platform by 12<br />
external collaborators.<br />
The Complex Portal contained over 1400 manually<br />
curated complexes at the end of <strong>2015</strong>. We launched<br />
an innovative graphical tool in the Complex Portal,<br />
which enables users to visualise complex topology and<br />
stoichiometry. This tool, originally developed by the<br />
Rapsilber group at the University of Edinburgh, was<br />
adapted by our developers to work with a new Java<br />
library (JAMI).<br />
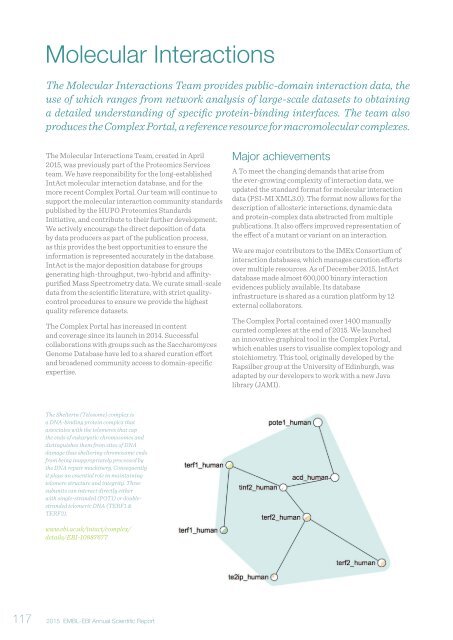
The Shelterin (Telosome) complex is<br />
a DNA-binding protein complex that<br />
associates with the telomeres that cap<br />
the ends of eukaryotic chromosomes and<br />
distinguishes them from sites of DNA<br />
damage thus sheltering chromosome ends<br />
from being inappropriately processed by<br />
the DNA repair machinery. Consequently<br />
it plays an essential role in maintaining<br />
telomere structure and integrity. Three<br />
subunits can interact directly either<br />
with single-stranded (POT1) or doublestranded<br />
telomeric DNA (TERF1 &<br />
TERF2).<br />
www.ebi.ac.uk/intact/complex/<br />
details/EBI-10887677<br />
117<br />
<strong>2015</strong> EMBL-EBI <strong>Annual</strong> <strong>Scientific</strong> <strong>Report</strong>