Annual Scientific Report 2015
EMBL_EBI_ASR_2015_DigitalEdition
EMBL_EBI_ASR_2015_DigitalEdition
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
PDBe Content and Integration<br />
PDBe aims to serve the biomedical community by providing easy access to<br />
macromolecular and cellular structure data. Our team is responsible for the<br />
curation of these data and for ensuring that the PDBe web resources serve the<br />
user community well. We also design tools to facilitate access to integrated,<br />
high-quality structural data.<br />
Major achievements<br />
Our major achievements in <strong>2015</strong> were completing the<br />
redesign of the PDBe website and annotating a record<br />
number of structures. In 2014, the wwPDB partners<br />
made the common deposition and annotation system<br />
available for X-ray crystal structures; as a consequence,<br />
the PDBe team has been handling all the European and<br />
African depositions to the archive, in addition to those<br />
made through the legacy deposition system. In addition,<br />
our curators annotate depositions to the EMDB archive,<br />
processing 4007 depositions to the PDB in <strong>2015</strong> (37%<br />
of 10 886 depositions worldwide) and annotating 452<br />
EMDB depositions (58% of 780 worldwide depositions).<br />
This is more than double the PDB depositions handled<br />
by the PDBe team in 2013. We managed this increase<br />
efficiently, returning more than 90% of the depositions<br />
to the depositors within two working days.<br />
As part of wwPDB development, we were involved<br />
in implementing the new common deposition and<br />
annotation system. This system provides, for the first<br />
time, a single interface for the deposition of data to the<br />
PDB and EMDB archives, and allows users to submit<br />
structures determined using X-ray, NMR and Electron<br />
Microscopy techniques. It was put into production in<br />
January 2016.<br />
The completion of the PDBe redesign project was<br />
a major milestone. It addressed the need to provide<br />
long-term sustainability through a robust weekly<br />
production process and improved management of<br />
PDBe infrastructure. It also put into effect an efficient<br />
data-discovery mechanism, providing intuitive,<br />
consistent data presentation based on user-centric<br />
approaches as a major step towards fulfilling our<br />
mission of ‘bringing structure to biology’.<br />
In 2014 and <strong>2015</strong>, we redesigned the<br />
PDBe infrastructure to support a<br />
robust weekly production process. We<br />
reworked the database infrastructure<br />
with help from the Systems<br />
Infrastructure team, and redesigned<br />
the web infrastructure with help from<br />
the Systems Applications and Web<br />
Production teams. The new weekly<br />
production pipeline has proven to be<br />
more robust and easier to maintain<br />
than the old system.<br />
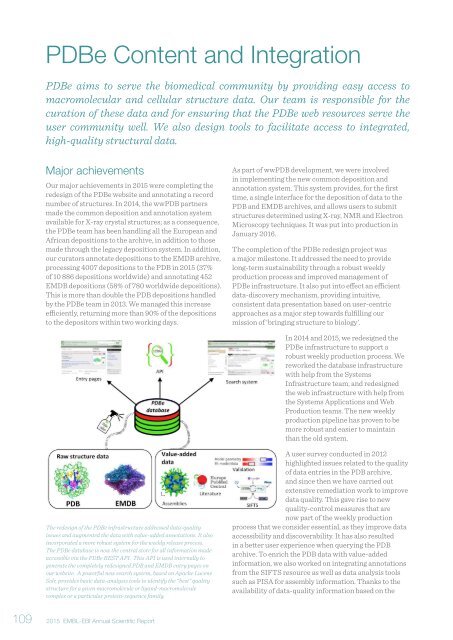
The redesign of the PDBe infrastructure addressed data-quality<br />
issues and augmented the data with value-added annotations. It also<br />
incorporated a more robust system for the weekly release process.<br />
The PDBe database is now the central store for all information made<br />
accessible via the PDBe REST API. This API is used internally to<br />
generate the completely redesigned PDB and EMDB entry pages on<br />
our website. A powerful new search system, based on Apache Lucene<br />
Solr, provides basic data-analysis tools to identify the “best” quality<br />
structure for a given macromolecule or ligand-macromolecule<br />
complex or a particular protein-sequence family.<br />
A user survey conducted in 2012<br />
highlighted issues related to the quality<br />
of data entries in the PDB archive,<br />
and since then we have carried out<br />
extensive remediation work to improve<br />
data quality. This gave rise to new<br />
quality-control measures that are<br />
now part of the weekly production<br />
process that we consider essential, as they improve data<br />
accessibility and discoverability. It has also resulted<br />
in a better user experience when querying the PDB<br />
archive. To enrich the PDB data with value-added<br />
information, we also worked on integrating annotations<br />
from the SIFTS resource as well as data analysis tools<br />
such as PISA for assembly information. Thanks to the<br />
availability of data-quality information based on the<br />
109<br />
<strong>2015</strong> EMBL-EBI <strong>Annual</strong> <strong>Scientific</strong> <strong>Report</strong>