Annual Scientific Report 2015
EMBL_EBI_ASR_2015_DigitalEdition
EMBL_EBI_ASR_2015_DigitalEdition
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
development of Europe PMC as a platform for<br />
application development based on full text. We<br />
contributed gene–disease associations text mined<br />
from both abstracts and open-access text articles to<br />
the Centre for Therapeutic Target Validation (Open<br />
Targets) platform, launched in December <strong>2015</strong>. For any<br />
given gene–disease association, articles are provided<br />
ranked by relevance. We now text mine 19 accession<br />
number types, adding resources such as the Gene<br />
Ontology, European Genome-phenome Archive (EGA)<br />
and ProteomeXchange to the list. This latter work of<br />
detecting data citations is particularly important to<br />
support literature-data integration, is contributing to<br />
our understanding of the impact of data resources in<br />
research, and is a key indicator for defining ELIXIR<br />
Core Resources.<br />
We now have 26 providers of external links from articles<br />
to websites of interest. This year we have added Publons,<br />
a small company that develops systems of credit for peer<br />
review, and Kudos, a company offering lay descriptions<br />
of research articles. The biggest supplier of links is now<br />
Wikipedia, with over 500 000 links to Wikipedia pages<br />
from articles in Europe PMC.<br />
Future plans<br />
In 2016 we plan to release a new system for displaying<br />
results from third-party application developers and<br />
text miners. Working with key communities across<br />
ELIXIR and beyond, we will develop the means to<br />
show text-mined annotations on open access articles<br />
on the Europe PMC website and share these results<br />
widely, for example to journals or other applications<br />
such as bibliographic reference managers. Using a<br />
Johanna McEntyre<br />
Literature Services<br />
PhD in plant biology, Manchester<br />
Metropolitan University, 1990. Editor,<br />
Trends in Biochemical Sciences, Elsevier,<br />
1997. Staff Scientist, NCBI, National Library<br />
of Medicine, NIH, US, 2009.<br />
At EMBL-EBI since 2009.<br />
variety of user-based research approaches, we will<br />
continue to improve the experience on the website, and<br />
in particular we expect to change the article pages and<br />
search experience for the better over the course of the<br />
year. We will be begin to support an author manuscript<br />
submission system, which has previously been hosted<br />
by our Jisc collaborators. A component of this is an<br />
Awarded Grants database containing all awards from<br />
the 27 Europe PMC funders since they joined. A new,<br />
integrated grant search will be released, showing the<br />
crossover between grants and the research outputs (in<br />
the form of articles) they funded.<br />
Selected publications<br />
Europe PMC Consortium (<strong>2015</strong>) Europe PMC: a<br />
full-text literature database for the life sciences<br />
and platform for innovation. Nucleic Acids Res.<br />
43:D1042-D1048<br />
Kafkas Ş, Kim JH, Pi X, McEntyre JR (<strong>2015</strong>) Database<br />
citation in supplementary data linked to Europe<br />
PubMed Central full text biomedical articles. J. Biomed.<br />
Semantics 6:1<br />
McEntyre J, Sarkans U, Brazma A (<strong>2015</strong>) The BioStudies<br />
database. Mol. Syst. Biol. 11:847<br />
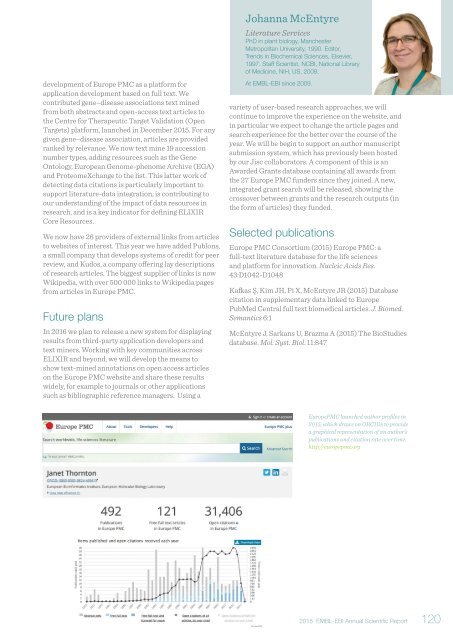
EuropePMC launched author profiles in<br />
<strong>2015</strong>, which draws on ORCIDs to provide<br />
a graphical representation of an author’s<br />
publications and citation rate over time.<br />
http://europepmc.org<br />
<strong>2015</strong> EMBL-EBI <strong>Annual</strong> <strong>Scientific</strong> <strong>Report</strong> 120