Annual Scientific Report 2015
EMBL_EBI_ASR_2015_DigitalEdition
EMBL_EBI_ASR_2015_DigitalEdition
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Daniel Zerbino<br />
Ensembl Genome Analysis<br />
MSc in Biotechnology, Ecole nationale supérieure<br />
des Mines de Paris, 2005. PhD in Bioinformatics,<br />
EMBL and University of Cambridge, 2009.<br />
At EMBL-EBI since 2013.<br />
Team leader since <strong>2015</strong>.<br />
<strong>2015</strong> to improve and automate our ChIP-Seq analysis<br />
pipelines, we will be processing most of the available<br />
datasets, as collected by IHEC. We will also integrate<br />
new data sources into Ensembl’s annotation. In<br />
particular, DNA methylation datasets are very cost<br />
effective, and many experiments have been done across<br />
many species. Currently only available on human and<br />
mouse, we are expecting to expand the number of species<br />
covered by the Ensembl Regulatory Build by integrating<br />
datasets produced by the FAANG consortium. In<br />
addition to better detecting regulatory elements, we<br />
wish to better understand their components, and<br />
transcription factor binding motifs in particular. New<br />
technologies such as SELEX or Uniprobe have produced<br />
new collections of binding motifs that we want to use in<br />
our detection of possible binding sites.<br />
Our annotation of variants will be greatly expanded<br />
by tying them empirically to nearby genes. First, we<br />
are very excited to release GTEx eQTL data in 2016.<br />
We have devoted a lot of efforts to develop appropriate<br />
HDF5-based infrastructure to store these large data<br />
matrices. Already, a prototype is running, and can<br />
provide any selection of the GTEx data in a fraction<br />
of a second. In parallel, we expect the first promoter<br />
capture Hi-C datasets to be made available around the<br />
same time. Our tissue specific annotations, as well as<br />
phenotype and disease associations, will all be enriched<br />
by the use of ontologies.<br />
pass, we will be displaying the CRISPR target sites, as<br />
computed by the WGE tool developed at the Sanger<br />
Institute. We will also be laying the groundwork to<br />
store external CRISPR screens in view of creating a<br />
central archive.<br />
In terms of tools we will continue to improve on our<br />
current set, with a focus on the VEP and GenomeStats.<br />
We are also currently collaborating with CTTV to<br />
produce a GWAS functional analysis pipeline, which<br />
will integrate all available cis-regulatory data to impute<br />
causal genes from GWAS summary results. This new<br />
service should be initially released in 2016, and will<br />
eventually be made available through various entry<br />
points, such as the Ensembl webpage, the RESTful<br />
endpoints or as a standalone program.<br />
Selected publications<br />
Cunningham F, et al. (<strong>2015</strong>) Improving the Sequence<br />
Ontology terminology for genomic variant annotation. J.<br />
Biomed. Semantics 6:32<br />
Nguyen N, et al. (<strong>2015</strong>) Building a pan-genome reference<br />
for a population. J. Comput. Biol. 22:387-401<br />
Zerbino DR, et al. (<strong>2015</strong>) The Ensembl regulatory build.<br />
Genome Biol. 16:56.<br />
We are also preparing for the fundamental revolution<br />
brought about by CRISPR technologies. In a first<br />
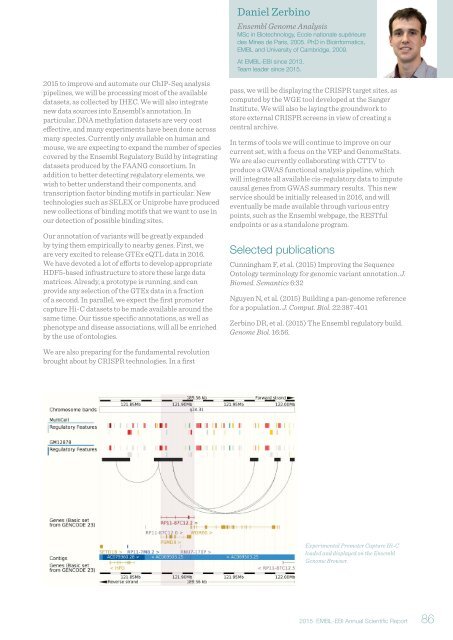
Experimental Promoter Capture Hi-C<br />
loaded and displayed on the Ensembl<br />
Genome Browser.<br />
<strong>2015</strong> EMBL-EBI <strong>Annual</strong> <strong>Scientific</strong> <strong>Report</strong> 86