Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Introduction<br />
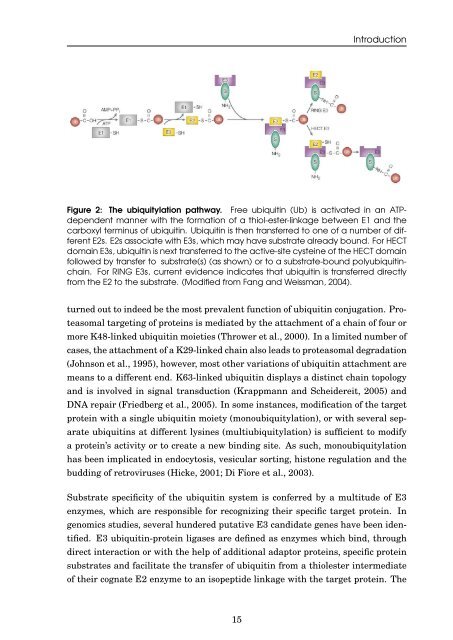
Figure 2: The ubiquitylation pathway. Free <strong>ubiquit<strong>in</strong></strong> (Ub) is activated <strong>in</strong> an ATPdependent<br />
manner with <strong>the</strong> formation <strong>of</strong> a thiol-ester-l<strong>in</strong>kage between E1 <strong>and</strong> <strong>the</strong><br />
carboxyl term<strong>in</strong>us <strong>of</strong> <strong>ubiquit<strong>in</strong></strong>. Ubiquit<strong>in</strong> is <strong>the</strong>n transferred to one <strong>of</strong> a number <strong>of</strong> different<br />
E2s. E2s associate with E3s, which may have substrate already bound. For HECT<br />
doma<strong>in</strong> E3s, <strong>ubiquit<strong>in</strong></strong> is next transferred to <strong>the</strong> active-site cyste<strong>in</strong>e <strong>of</strong> <strong>the</strong> HECT doma<strong>in</strong><br />
followed by transfer to substrate(s) (as shown) or to a substrate-bound poly<strong>ubiquit<strong>in</strong></strong>cha<strong>in</strong>.<br />
For RING E3s, current evidence <strong>in</strong>dicates that <strong>ubiquit<strong>in</strong></strong> is transferred directly<br />
from <strong>the</strong> E2 to <strong>the</strong> substrate. (Modified from Fang <strong>and</strong> Weissman, 2004).<br />
turned out to <strong>in</strong>deed be <strong>the</strong> most prevalent function <strong>of</strong> <strong>ubiquit<strong>in</strong></strong> conjugation. Pro-<br />
teasomal target<strong>in</strong>g <strong>of</strong> prote<strong>in</strong>s is mediated by <strong>the</strong> attachment <strong>of</strong> a cha<strong>in</strong> <strong>of</strong> four or<br />
more K48-l<strong>in</strong>ked <strong>ubiquit<strong>in</strong></strong> moieties (Thrower et al., 2000). In a limited number <strong>of</strong><br />
cases, <strong>the</strong> attachment <strong>of</strong> a K29-l<strong>in</strong>ked cha<strong>in</strong> also leads to proteasomal <strong>degradation</strong><br />
(Johnson et al., 1995), however, most o<strong>the</strong>r variations <strong>of</strong> <strong>ubiquit<strong>in</strong></strong> attachment are<br />
means to a different end. K63-l<strong>in</strong>ked <strong>ubiquit<strong>in</strong></strong> displays a dist<strong>in</strong>ct cha<strong>in</strong> topology<br />
<strong>and</strong> is <strong>in</strong>volved <strong>in</strong> signal transduction (Krappmann <strong>and</strong> Scheidereit, 2005) <strong>and</strong><br />
DNA repair (Friedberg et al., 2005). In some <strong>in</strong>stances, modification <strong>of</strong> <strong>the</strong> target<br />
prote<strong>in</strong> with a s<strong>in</strong>gle <strong>ubiquit<strong>in</strong></strong> moiety (monoubiquitylation), or with several sep-<br />
arate <strong>ubiquit<strong>in</strong></strong>s at different lys<strong>in</strong>es (multiubiquitylation) is sufficient to modify<br />
a prote<strong>in</strong>’s activity or to create a new b<strong>in</strong>d<strong>in</strong>g site. As such, monoubiquitylation<br />
has been implicated <strong>in</strong> endocytosis, vesicular sort<strong>in</strong>g, histone regulation <strong>and</strong> <strong>the</strong><br />
budd<strong>in</strong>g <strong>of</strong> retroviruses (Hicke, 2001; Di Fiore et al., 2003).<br />
Substrate specificity <strong>of</strong> <strong>the</strong> <strong>ubiquit<strong>in</strong></strong> system is conferred by a multitude <strong>of</strong> E3<br />
enzymes, which are responsible for recogniz<strong>in</strong>g <strong>the</strong>ir specific target prote<strong>in</strong>. In<br />
genomics studies, several hundered putative E3 c<strong>and</strong>idate genes have been iden-<br />
tified. E3 <strong>ubiquit<strong>in</strong></strong>-prote<strong>in</strong> ligases are def<strong>in</strong>ed as enzymes which b<strong>in</strong>d, through<br />
direct <strong>in</strong>teraction or with <strong>the</strong> help <strong>of</strong> additional adaptor prote<strong>in</strong>s, specific prote<strong>in</strong><br />
substrates <strong>and</strong> facilitate <strong>the</strong> transfer <strong>of</strong> <strong>ubiquit<strong>in</strong></strong> from a thiolester <strong>in</strong>termediate<br />
<strong>of</strong> <strong>the</strong>ir cognate E2 enzyme to an isopeptide l<strong>in</strong>kage with <strong>the</strong> target prote<strong>in</strong>. The<br />
15