Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Chapter 1<br />
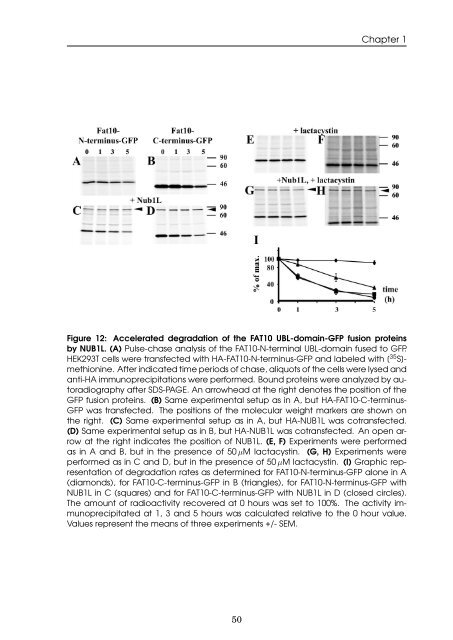
Figure 12: Accelerated <strong>degradation</strong> <strong>of</strong> <strong>the</strong> <strong>FAT10</strong> UBL-doma<strong>in</strong>-GFP fusion prote<strong>in</strong>s<br />
by NUB1L. (A) Pulse-chase analysis <strong>of</strong> <strong>the</strong> <strong>FAT10</strong>-N-term<strong>in</strong>al UBL-doma<strong>in</strong> fused to GFP.<br />
HEK293T cells were transfected with HA-<strong>FAT10</strong>-N-term<strong>in</strong>us-GFP <strong>and</strong> labeled with [ 35 S]methion<strong>in</strong>e.<br />
After <strong>in</strong>dicated time periods <strong>of</strong> chase, aliquots <strong>of</strong> <strong>the</strong> cells were lysed <strong>and</strong><br />
anti-HA immunoprecipitations were performed. Bound prote<strong>in</strong>s were analyzed by autoradiography<br />
after SDS-PAGE. An arrowhead at <strong>the</strong> right denotes <strong>the</strong> position <strong>of</strong> <strong>the</strong><br />
GFP fusion prote<strong>in</strong>s. (B) Same experimental setup as <strong>in</strong> A, but HA-<strong>FAT10</strong>-C-term<strong>in</strong>us-<br />
GFP was transfected. The positions <strong>of</strong> <strong>the</strong> molecular weight markers are shown on<br />
<strong>the</strong> right. (C) Same experimental setup as <strong>in</strong> A, but HA-NUB1L was cotransfected.<br />
(D) Same experimental setup as <strong>in</strong> B, but HA-NUB1L was cotransfected. An open arrow<br />
at <strong>the</strong> right <strong>in</strong>dicates <strong>the</strong> position <strong>of</strong> NUB1L. (E, F) Experiments were performed<br />
as <strong>in</strong> A <strong>and</strong> B, but <strong>in</strong> <strong>the</strong> presence <strong>of</strong> 50 µM lactacyst<strong>in</strong>. (G, H) Experiments were<br />
performed as <strong>in</strong> C <strong>and</strong> D, but <strong>in</strong> <strong>the</strong> presence <strong>of</strong> 50 µM lactacyst<strong>in</strong>. (I) Graphic representation<br />
<strong>of</strong> <strong>degradation</strong> rates as determ<strong>in</strong>ed for <strong>FAT10</strong>-N-term<strong>in</strong>us-GFP alone <strong>in</strong> A<br />
(diamonds), for <strong>FAT10</strong>-C-term<strong>in</strong>us-GFP <strong>in</strong> B (triangles), for <strong>FAT10</strong>-N-term<strong>in</strong>us-GFP with<br />
NUB1L <strong>in</strong> C (squares) <strong>and</strong> for <strong>FAT10</strong>-C-term<strong>in</strong>us-GFP with NUB1L <strong>in</strong> D (closed circles).<br />
The amount <strong>of</strong> radioactivity recovered at 0 hours was set to 100%. The activity immunoprecipitated<br />
at 1, 3 <strong>and</strong> 5 hours was calculated relative to <strong>the</strong> 0 hour value.<br />
Values represent <strong>the</strong> means <strong>of</strong> three experiments +/- SEM.<br />
50