Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Chapter 3<br />
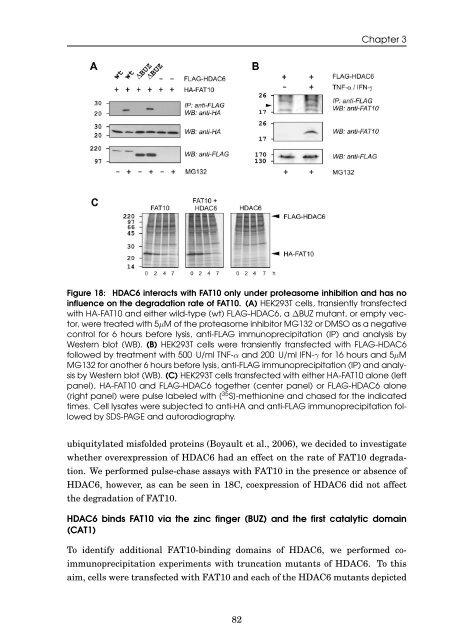
Figure 18: HDAC6 <strong>in</strong>teracts with <strong>FAT10</strong> only under proteasome <strong>in</strong>hibition <strong>and</strong> has no<br />
<strong>in</strong>fluence on <strong>the</strong> <strong>degradation</strong> rate <strong>of</strong> <strong>FAT10</strong>. (A) HEK293T cells, transiently transfected<br />
with HA-<strong>FAT10</strong> <strong>and</strong> ei<strong>the</strong>r wild-type (wt) FLAG-HDAC6, a ∆BUZ mutant, or empty vector,<br />
were treated with 5µM <strong>of</strong> <strong>the</strong> proteasome <strong>in</strong>hibitor MG132 or DMSO as a negative<br />
control for 6 hours before lysis, anti-FLAG immunoprecipitation (IP) <strong>and</strong> analysis by<br />
Western blot (WB). (B) HEK293T cells were transiently transfected with FLAG-HDAC6<br />
followed by treatment with 500 U/ml TNF-α <strong>and</strong> 200 U/ml IFN-γ for 16 hours <strong>and</strong> 5µM<br />
MG132 for ano<strong>the</strong>r 6 hours before lysis, anti-FLAG immunoprecipitation (IP) <strong>and</strong> analysis<br />
by Western blot (WB). (C) HEK293T cells transfected with ei<strong>the</strong>r HA-<strong>FAT10</strong> alone (left<br />
panel), HA-<strong>FAT10</strong> <strong>and</strong> FLAG-HDAC6 toge<strong>the</strong>r (center panel) or FLAG-HDAC6 alone<br />
(right panel) were pulse labeled with [ 35 S]-methion<strong>in</strong>e <strong>and</strong> chased for <strong>the</strong> <strong>in</strong>dicated<br />
times. Cell lysates were subjected to anti-HA <strong>and</strong> anti-FLAG immunoprecipitation followed<br />
by SDS-PAGE <strong>and</strong> autoradiography.<br />
ubiquitylated misfolded prote<strong>in</strong>s (Boyault et al., 2006), we decided to <strong>in</strong>vestigate<br />
whe<strong>the</strong>r overexpression <strong>of</strong> HDAC6 had an effect on <strong>the</strong> rate <strong>of</strong> <strong>FAT10</strong> degrada-<br />
tion. We performed pulse-chase assays with <strong>FAT10</strong> <strong>in</strong> <strong>the</strong> presence or absence <strong>of</strong><br />
HDAC6, however, as can be seen <strong>in</strong> 18C, coexpression <strong>of</strong> HDAC6 did not affect<br />
<strong>the</strong> <strong>degradation</strong> <strong>of</strong> <strong>FAT10</strong>.<br />
HDAC6 b<strong>in</strong>ds <strong>FAT10</strong> via <strong>the</strong> z<strong>in</strong>c f<strong>in</strong>ger (BUZ) <strong>and</strong> <strong>the</strong> first catalytic doma<strong>in</strong><br />
(CAT1)<br />
To identify additional <strong>FAT10</strong>-b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong>s <strong>of</strong> HDAC6, we performed co-<br />
immunoprecipitation experiments with truncation mutants <strong>of</strong> HDAC6. To this<br />
aim, cells were transfected with <strong>FAT10</strong> <strong>and</strong> each <strong>of</strong> <strong>the</strong> HDAC6 mutants depicted<br />
82