Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
Role of the ubiquitin-like modifier FAT10 in protein degradation and ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Chapter 2<br />
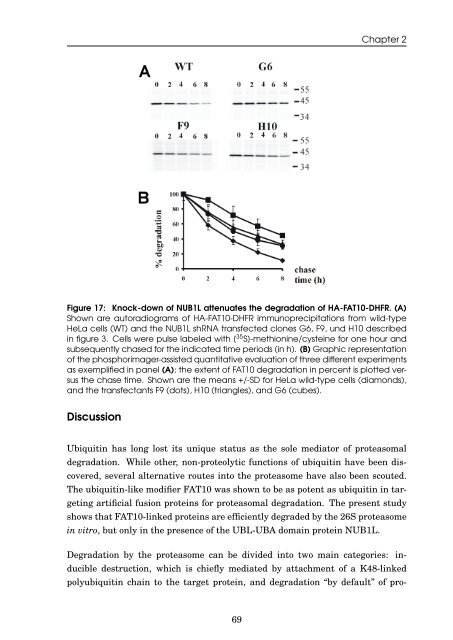
Figure 17: Knock-down <strong>of</strong> NUB1L attenuates <strong>the</strong> <strong>degradation</strong> <strong>of</strong> HA-<strong>FAT10</strong>-DHFR. (A)<br />
Shown are autoradiograms <strong>of</strong> HA-<strong>FAT10</strong>-DHFR immunoprecipitations from wild-type<br />
HeLa cells (WT) <strong>and</strong> <strong>the</strong> NUB1L shRNA transfected clones G6, F9, und H10 described<br />
<strong>in</strong> figure 3. Cells were pulse labeled with [ 35 S]-methion<strong>in</strong>e/cyste<strong>in</strong>e for one hour <strong>and</strong><br />
subsequently chased for <strong>the</strong> <strong>in</strong>dicated time periods (<strong>in</strong> h). (B) Graphic representation<br />
<strong>of</strong> <strong>the</strong> phosphorimager-assisted quantitative evaluation <strong>of</strong> three different experiments<br />
as exemplified <strong>in</strong> panel (A); <strong>the</strong> extent <strong>of</strong> <strong>FAT10</strong> <strong>degradation</strong> <strong>in</strong> percent is plotted versus<br />
<strong>the</strong> chase time. Shown are <strong>the</strong> means +/-SD for HeLa wild-type cells (diamonds),<br />
<strong>and</strong> <strong>the</strong> transfectants F9 (dots), H10 (triangles), <strong>and</strong> G6 (cubes).<br />
Discussion<br />
Ubiquit<strong>in</strong> has long lost its unique status as <strong>the</strong> sole mediator <strong>of</strong> proteasomal<br />
<strong>degradation</strong>. While o<strong>the</strong>r, non-proteolytic functions <strong>of</strong> <strong>ubiquit<strong>in</strong></strong> have been dis-<br />
covered, several alternative routes <strong>in</strong>to <strong>the</strong> proteasome have also been scouted.<br />
The <strong>ubiquit<strong>in</strong></strong>-<strong>like</strong> <strong>modifier</strong> <strong>FAT10</strong> was shown to be as potent as <strong>ubiquit<strong>in</strong></strong> <strong>in</strong> tar-<br />
get<strong>in</strong>g artificial fusion prote<strong>in</strong>s for proteasomal <strong>degradation</strong>. The present study<br />
shows that <strong>FAT10</strong>-l<strong>in</strong>ked prote<strong>in</strong>s are efficiently degraded by <strong>the</strong> 26S proteasome<br />
<strong>in</strong> vitro, but only <strong>in</strong> <strong>the</strong> presence <strong>of</strong> <strong>the</strong> UBL-UBA doma<strong>in</strong> prote<strong>in</strong> NUB1L.<br />
Degradation by <strong>the</strong> proteasome can be divided <strong>in</strong>to two ma<strong>in</strong> categories: <strong>in</strong>-<br />
ducible destruction, which is chiefly mediated by attachment <strong>of</strong> a K48-l<strong>in</strong>ked<br />
poly<strong>ubiquit<strong>in</strong></strong> cha<strong>in</strong> to <strong>the</strong> target prote<strong>in</strong>, <strong>and</strong> <strong>degradation</strong> “by default” <strong>of</strong> pro-<br />
69