biologia - Studia
biologia - Studia
biologia - Studia
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
STRUCTURE AND DYNAMICS OF THE PREDATOR MITE’S POPULATIONS IN SHRUBS ECOSYSTEMS<br />
2500<br />
2000<br />
Lunca Mare<br />
Cornu<br />
Nistoresti<br />
no. ind./sq.m.<br />
1500<br />
1000<br />
500<br />
0<br />
Leptogamasus parvulus<br />
Veigaia nemorensis<br />
Asca aphidoides<br />
Geholaspis mandibularis<br />
Hypoaspis aculeifer<br />
species<br />
Trachytes aegrota<br />
Uropoda sp.<br />
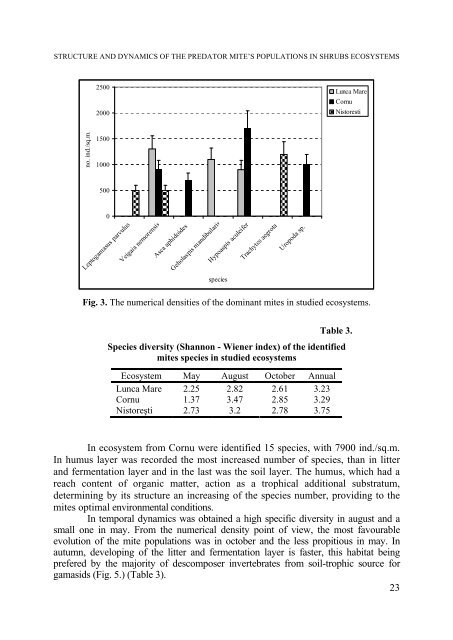
Fig. 3. The numerical densities of the dominant mites in studied ecosystems.<br />
Table 3.<br />
Species diversity (Shannon - Wiener index) of the identified<br />
mites species in studied ecosystems<br />
Ecosystem May August October Annual<br />
Lunca Mare 2.25 2.82 2.61 3.23<br />
Cornu 1.37 3.47 2.85 3.29<br />
Nistoreşti 2.73 3.2 2.78 3.75<br />
In ecosystem from Cornu were identified 15 species, with 7900 ind./sq.m.<br />
In humus layer was recorded the most increased number of species, than in litter<br />
and fermentation layer and in the last was the soil layer. The humus, which had a<br />
reach content of organic matter, action as a trophical additional substratum,<br />
determining by its structure an increasing of the species number, providing to the<br />
mites optimal environmental conditions.<br />
In temporal dynamics was obtained a high specific diversity in august and a<br />
small one in may. From the numerical density point of view, the most favourable<br />
evolution of the mite populations was in october and the less propitious in may. In<br />
autumn, developing of the litter and fermentation layer is faster, this habitat being<br />
prefered by the majority of descomposer invertebrates from soil-trophic source for<br />
gamasids (Fig. 5.) (Table 3).<br />
23