H e m a t o lo g y E d u c a t io n - European Hematology Association
H e m a t o lo g y E d u c a t io n - European Hematology Association
H e m a t o lo g y E d u c a t io n - European Hematology Association
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
of cases) 46 and amplificat<strong>io</strong>ns of the reg<strong>io</strong>n encompassing<br />
the mir-17-92 microRNA cluster on chromosome 13,<br />
which has also been shown to suppress PTEN. 60<br />
ABC-DLBCL<br />
Several genetic abnormalities are observed almost<br />
exclusively in ABC-DLBCL, including amplificat<strong>io</strong>ns of<br />
the BCL2 <strong>lo</strong>cus on 18q24; 61,62 mutat<strong>io</strong>ns within the NFκB<br />
(CARD11, TNFAIP3/A20), 63,64 BCR (CD79A/B) 65 and<br />
JAK/STAT (MYD88) signaling pathways; inactivating<br />
mutat<strong>io</strong>ns and delet<strong>io</strong>ns of PRDM1; 27,28,66 and delet<strong>io</strong>n or<br />
lack of express<strong>io</strong>n of the CDKN2A tumor suppressor<br />
gene. In addit<strong>io</strong>n, chromosomal trans<strong>lo</strong>cat<strong>io</strong>ns deregulating<br />
the BCL6 oncogene are found more frequently in<br />
this subtype. Mutat<strong>io</strong>ns of ATM have also been reported<br />
in a small subset of cases. 67,68<br />
NF-κB pathway les<strong>io</strong>ns<br />
A prominent feature of ABC-DLBCL is the constitutive<br />
activat<strong>io</strong>n of the NF-κB signaling pathway, as<br />
revealed by the selective enrichment in NF-κB target<br />
genes express<strong>io</strong>n, and by the requirement of NF-κB for<br />
the proliferat<strong>io</strong>n and survival of ABC-, but not GCB-<br />
DLBCL cell lines. 85 Over the past two years, a number of<br />
studies have provided a genetic basis for this phenotypic<br />
characteristic by identifying multiple oncogenic alterat<strong>io</strong>ns<br />
that affect positive and negative regulators of NFκB,<br />
specifically in this disease subtype. In up to 30% of<br />
the cases, biallelic mutat<strong>io</strong>ns and/or delet<strong>io</strong>ns inactivate<br />
the TNFAIP3 gene, which encodes for the negative regulator<br />
A20, thus preventing terminat<strong>io</strong>n of NF-κB<br />
responses. 63,86 In ~9% of patients, CARD11 is targeted<br />
by oncogenic mutat<strong>io</strong>ns that cluster within the protein<br />
coiled-coil domain and enhance its ability to transactivate<br />
NF-κB target genes. 63,64 Less commonly, mutat<strong>io</strong>ns<br />
were found in a variety of other genes encoding for NF-κB<br />
components (among others, TRAF2 and TRAF5). 63<br />
London, United Kingdom, June 9-12, 2011<br />
ABC-DLBCL activate NF-κB also via a chronic form of<br />
active BCR signaling, which is associated with somatic<br />
mutat<strong>io</strong>ns in the immunoreceptor tyrosine-based activat<strong>io</strong>n<br />
motif (ITAM) signaling modules of CD79B and<br />
CD79A (> 20% of patients). 65 Since BCR signaling can<br />
also trigger other pathways, such as MAPK and PI3K,<br />
future studies will have to address the relative or coordinate<br />
contribut<strong>io</strong>n of these pathways to the deve<strong>lo</strong>pment<br />
of DLBCL (Figure 2). Finally, oncogenically active<br />
MYD88 mutat<strong>io</strong>ns were recently reported in almost one<br />
third of all ABC-DLBCLs, where they mostly target an<br />
evolut<strong>io</strong>narily invariant residue within the TIR (Toll/IL1<br />
receptor) domain. 87 Besides activating NF-κB, mutat<strong>io</strong>ns<br />
of MYD88 induce JAK/STAT3 transcript<strong>io</strong>nal responses,<br />
also a phenotypic trait of ABC-DLBCL. 88,89 Collectively,<br />
les<strong>io</strong>ns converging on the NF-κB pathway account for<br />
greater than 50% of all ABC-DLBCL. 63<br />
PRDM1 inactivat<strong>io</strong>n<br />
In up to 25% of ABC-DLBCLs, the PRDM1 gene is<br />
inactivated by a variety of genetic les<strong>io</strong>ns, including<br />
truncating point mutat<strong>io</strong>ns, inactivating missense mutat<strong>io</strong>ns,<br />
and/or genomic delet<strong>io</strong>ns. 27,28,66 An addit<strong>io</strong>nal sizeable<br />
fract<strong>io</strong>n of cases has <strong>lo</strong>st PRDM1 protein express<strong>io</strong>n<br />
due to transcript<strong>io</strong>nal repress<strong>io</strong>n by constitutively<br />
active, trans<strong>lo</strong>cated BCL6 alleles. 27,66 The PRDM1 gene<br />
encodes for a zinc finger transcript<strong>io</strong>nal repressor that is<br />
expressed in a subset of GC B cells undergoing plasma<br />
cell differentiat<strong>io</strong>n and in all plasma cells, 82,83 and which<br />
is essential for terminal B cell differentiat<strong>io</strong>n. 84 Thus,<br />
PRDM1 funct<strong>io</strong>ns as a tumor suppressor gene and may<br />
favor malignant transformat<strong>io</strong>n by b<strong>lo</strong>cking post-GC<br />
differentiat<strong>io</strong>n of B cells. In line with these findings,<br />
rearrangements of the BCL6 <strong>lo</strong>cus and genetic les<strong>io</strong>ns of<br />
PRDM1 are mutually exclusive in ABC-DLBCL, indicating<br />
that BCL6 deregulat<strong>io</strong>n and PRDM1 inactivat<strong>io</strong>n<br />
represent alternative oncogenic mechanisms converging<br />
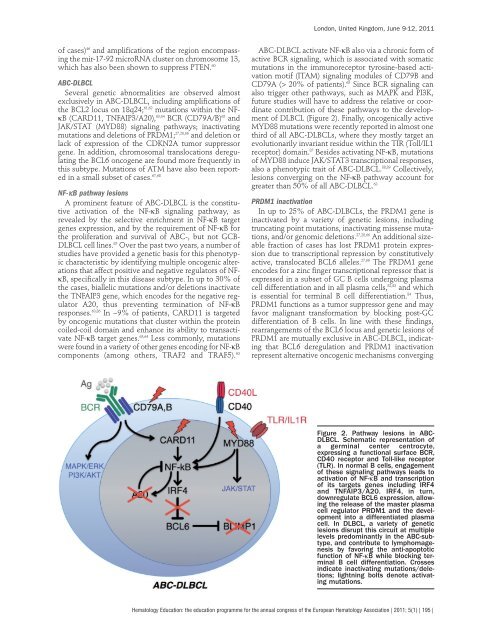
Figure 2. Pathway les<strong>io</strong>ns in ABC-<br />
DLBCL. Schematic representat<strong>io</strong>n of<br />
a germinal center centrocyte,<br />
expressing a funct<strong>io</strong>nal surface BCR,<br />
CD40 receptor and Toll-like receptor<br />
(TLR). In normal B cells, engagement<br />
of these signaling pathways leads to<br />
activat<strong>io</strong>n of NF-κB and transcript<strong>io</strong>n<br />
of its targets genes including IRF4<br />
and TNFAIP3/A20. IRF4, in turn,<br />
downregulate BCL6 express<strong>io</strong>n, al<strong>lo</strong>wing<br />
the release of the master plasma<br />
cell regulator PRDM1 and the deve<strong>lo</strong>pment<br />
into a differentiated plasma<br />
cell. In DLBCL, a variety of genetic<br />
les<strong>io</strong>ns disrupt this circuit at multiple<br />
levels predominantly in the ABC-subtype,<br />
and contribute to lymphomagenesis<br />
by favoring the anti-apoptotic<br />
funct<strong>io</strong>n of NF-κB while b<strong>lo</strong>cking terminal<br />
B cell differentiat<strong>io</strong>n. Crosses<br />
indicate inactivating mutat<strong>io</strong>ns/delet<strong>io</strong>ns;<br />
lightning bolts denote activating<br />
mutat<strong>io</strong>ns.<br />
Hemato<strong>lo</strong>gy Educat<strong>io</strong>n: the educat<strong>io</strong>n programme for the annual congress of the <strong>European</strong> Hemato<strong>lo</strong>gy Associat<strong>io</strong>n | 2011; 5(1) | 195 |