H e m a t o lo g y E d u c a t io n - European Hematology Association
H e m a t o lo g y E d u c a t io n - European Hematology Association
H e m a t o lo g y E d u c a t io n - European Hematology Association
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
16 th Congress of the <strong>European</strong> Hemato<strong>lo</strong>gy Associat<strong>io</strong>n<br />
either a truncated dominant negative isoform or protein<br />
with decreased DNA binding or dimerizat<strong>io</strong>n ability.<br />
Approximately 85% of CEBPA mutant AMLs are biallelic<br />
so that no wild type protein is produced, 12 and these<br />
patients have a particularly good prognosis. 13 By contrast,<br />
mutat<strong>io</strong>n of a single allele of CEBPA does not<br />
affect prognosis. NRAS (9–14%) and KRAS (5–17%)<br />
mutat<strong>io</strong>ns are of uncertain prognostic significance. In<br />
NK-AML, overexpress<strong>io</strong>n of the BAALC, ERG, MN1,<br />
and EVI1 genes is associated with poor prognosis. 14,15<br />
Poor prognosis AML includes patients with complex<br />
karyotype (harboring at least four unrelated cytogenetic<br />
abnormalities). 16 These patients tend to be older and<br />
may have an antecedent mye<strong>lo</strong>dysplastic or mye<strong>lo</strong>proliferative<br />
disorder. Mutat<strong>io</strong>ns involving the TP53 occur<br />
frequently in this group (56–78%). 17 Amongst the<br />
adverse prognosis group, multiple studies showed that<br />
monosomal karyotype (MK), defined as autosomal<br />
monosomy in the presence of another autosomal monosomy<br />
or other chromosomal abnormalities, is associated<br />
with a particularly poor prognosis, with overall survival<br />
rates of less than 5%. 16,18,19<br />
Single nucleotide polymorphism (SNP) arrays, which<br />
can identify small genomic amplificat<strong>io</strong>ns, delet<strong>io</strong>ns,<br />
and areas of copy number neutral <strong>lo</strong>ss of heterozygosity<br />
(LOH), also known as uniparental disomy (UPD),<br />
showed that approximately 20% of NK-AML exhibited<br />
partial UPD. 20 Later studies revealed that AML is characterized<br />
by a high number of small, non-recurrent copy<br />
number alterat<strong>io</strong>ns. 21 SNP arrays identified TET2 as a<br />
potential tumor suppressor in AML and other mye<strong>lo</strong>id<br />
neoplasms, as TET2 mutat<strong>io</strong>ns are found in 24% of secondary<br />
AML, 19% of mye<strong>lo</strong>dysplastic syndrome<br />
(MDS), 12% of mye<strong>lo</strong>proliferative neoplasms (MPN),<br />
and 22% of chronic mye<strong>lo</strong>monocytic leukemia<br />
(CMML). 22 Over the past 2 years, whole-genome<br />
sequencing discovered recurrent mutat<strong>io</strong>ns in AML,<br />
such as DNMT3A 23 and IDH1. 24 DNMT3A mutat<strong>io</strong>ns<br />
were enriched in patients with intermediate risk cytoge-<br />
netics (33.7%) and conferred a poor prognosis. IDH1<br />
and IDH2 mutat<strong>io</strong>ns were found in 14% and 19% of<br />
NK-AML, respectively and the impact of these mutat<strong>io</strong>ns<br />
on outcome is uncertain and may be influenced by<br />
the presence of coincident mutat<strong>io</strong>ns (reviewed by<br />
Lowenberg B. in this issue). 25,26 Large-scale sequencing<br />
also revealed mutat<strong>io</strong>ns in ASXL1 in 6% and 53% of<br />
patients with primary and secondary AML. 27<br />
AML may be classified by gene express<strong>io</strong>n, DNA<br />
methylat<strong>io</strong>n, and chromatin modificat<strong>io</strong>n patterns.<br />
Gene express<strong>io</strong>n profiling can predict FAB subtypes 28<br />
and discern novel prognostic subsets of AML. 29–31 The<br />
combinat<strong>io</strong>n of gene express<strong>io</strong>n and promoter methylat<strong>io</strong>n<br />
signatures, in which the transcript<strong>io</strong>nal capacity of<br />
the genome is ascertained, can further identify AML<br />
subtypes. 32 Figueroa et al. found 16 different patterns of<br />
promoter methylat<strong>io</strong>n among AML. While some corresponded<br />
to known cytogenetically defined subtypes,<br />
there were five methylat<strong>io</strong>n-defined AML subtypes<br />
with no other defined genetic characteristics. Several of<br />
these groups were subsequently shown to correspond<br />
to patients with TET2 or IDH1 or IDH2 mutat<strong>io</strong>ns. 33<br />
The methylat<strong>io</strong>n status of 15 promoters could be used<br />
as a predictor of survival. Analysis of histone methylat<strong>io</strong>n<br />
patterns in primary AML specimens may also be<br />
useful. Profiling of H3K9 promoter methylat<strong>io</strong>n<br />
revealed distinct differences between AML blasts and<br />
normal CD34+ cells and found that the state of H3K9<br />
methylat<strong>io</strong>n was a predictor of prognosis. 34 Unlike DNA<br />
methylat<strong>io</strong>n profiling, however, H3K9 methylat<strong>io</strong>n patterns<br />
were not associated with known cytogenetic<br />
abnormalities and were not correlated with gene<br />
express<strong>io</strong>n.<br />
MicroRNAs play an important role in normal<br />
hematopoiesis and leukemogenesis. 35 Genome-wide<br />
microRNA analysis in AML identified a correlat<strong>io</strong>n<br />
between microRNA signatures, specific cytogenetic<br />
groups, and prognosis. 36,37 In NK-AML, a microRNA<br />
signature was identified for high-risk patients harbor-<br />
| 28 | Hemato<strong>lo</strong>gy Educat<strong>io</strong>n: the educat<strong>io</strong>n programme for the annual congress of the <strong>European</strong> Hemato<strong>lo</strong>gy Associat<strong>io</strong>n | 2011; 5(1)<br />
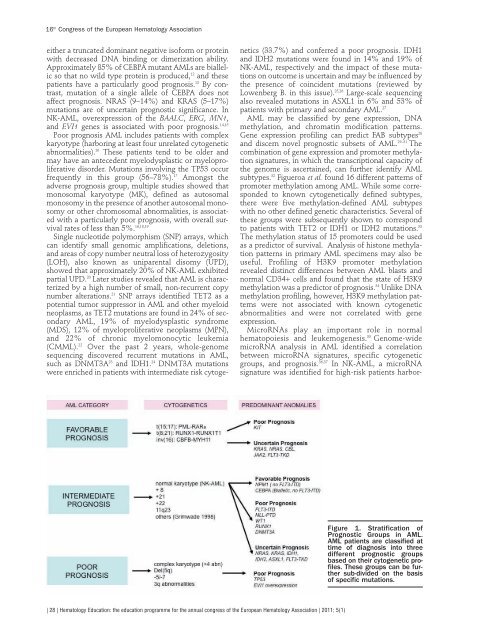
Figure 1. Stratificat<strong>io</strong>n of<br />
Prognostic Groups in AML.<br />
AML patients are classified at<br />
time of diagnosis into three<br />
different prognostic groups<br />
based on their cytogenetic profiles.<br />
These groups can be further<br />
sub-divided on the basis<br />
of specific mutat<strong>io</strong>ns.