et al.
et al.
et al.
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
of the MS spectra reve<strong>al</strong>ed two ions with massto-charge<br />
ratios (m/z) of 675.1 (z =1 + ) and 338.1<br />
(z =2 + ), which were present in the active fractions<br />
but absent in the background spectra (Fig. 2A).<br />
These m/z v<strong>al</strong>ues, despite the low mass accuracy<br />
of the mass spectrom<strong>et</strong>er (LTQ, Thermo), were<br />
equiv<strong>al</strong>ent to the average c<strong>al</strong>culated m/z v<strong>al</strong>ues of<br />
c-di-GMP and c-di-AMP [675 = (691 + 659)/2].<br />
This observation suggested that the d<strong>et</strong>ected ion<br />
was a hybrid of c-di-GMP and c-di-AMP—that<br />
is, cyclic GMP-AMP, or cGAMP (m/z = 675.107,<br />
z =1 + ; m/z = 338.057, z =2 + ). Collision-induced<br />
dissociation (CID) fragmentation of this ion (m/z =<br />
338.1, z =2 + ) reve<strong>al</strong>ed sever<strong>al</strong> prominent ions<br />
with m/z v<strong>al</strong>ues expected of the product ions of<br />
cGAMP (Fig. 2B). Quantitative mass spectrom<strong>et</strong>ry<br />
using selective reaction monitoring (SRM)<br />
showed that the abundance of the ions representing<br />
cGAMP in the fractions from a C18 column<br />
correlated very well with their IRF3-stimulatory<br />
activities (Fig. 2C and fig. S2C). cGAMP has recently<br />
been identified in the bacterium Vibrio<br />
cholerae and shown to play a role in bacteri<strong>al</strong> chemotaxis<br />
and colonization (10). However, cGAMP<br />
has not been reported to exist or function in eukaryotic<br />
cells.<br />
To verify the identity of the heat-resistant<br />
STING activator, we used a high-resolution highaccuracy<br />
mass spectrom<strong>et</strong>er (Q Exactive, Thermo)<br />
to perform nano-LC-MS an<strong>al</strong>ysis. The cell-derived<br />
STING activator had m/z v<strong>al</strong>ues of 675.107 (z =1 + )<br />
and 338.057 (z =2 + ), which exactly matched<br />
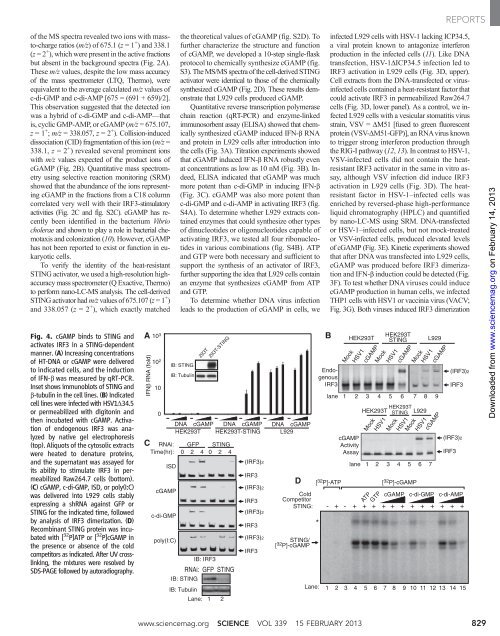
Fig. 4. cGAMP binds to STING and<br />
activates IRF3 in a STING-dependent<br />
manner. (A) Increasing concentrations<br />
of HT-DNA or cGAMP were delivered<br />
to indicated cells, and the induction<br />
of IFN-b was measured by qRT-PCR.<br />
Ins<strong>et</strong> shows immunoblots of STING and<br />
b-tubulin in the cell lines. (B)Indicated<br />
cell lines were infected with HSV1D34.5<br />
or permeabilized with digitonin and<br />
then incubated with cGAMP. Activation<br />
of endogenous IRF3 was an<strong>al</strong>yzed<br />
by native gel electrophoresis<br />
(top). Aliquots of the cytosolic extracts<br />
were heated to denature proteins,<br />
and the supernatant was assayed for<br />
its ability to stimulate IRF3 in permeabilized<br />
Raw264.7 cells (bottom).<br />
(C) cGAMP, c-di-GMP, ISD, or poly(I:C)<br />
was delivered into L929 cells stably<br />
expressing a shRNA against GFP or<br />
STING for the indicated time, followed<br />
by an<strong>al</strong>ysis of IRF3 dimerization. (D)<br />
Recombinant STING protein was incubated<br />
with [ 32 P]ATP or [ 32 P]cGAMP in<br />
the presence or absence of the cold<br />
comp<strong>et</strong>itors as indicated. After UV crosslinking,<br />
the mixtures were resolved by<br />
SDS-PAGE followed by autoradiography.<br />
the theor<strong>et</strong>ic<strong>al</strong> v<strong>al</strong>ues of cGAMP (fig. S2D). To<br />
further characterize the structure and function<br />
of cGAMP, we developed a 10-step single-flask<br />
protocol to chemic<strong>al</strong>ly synthesize cGAMP (fig.<br />
S3). The MS/MS spectra of the cell-derived STING<br />
activator were identic<strong>al</strong> to those of the chemic<strong>al</strong>ly<br />
synthesized cGAMP (Fig. 2D). These results demonstrate<br />
that L929 cells produced cGAMP.<br />
Quantitative reverse transcription polymerase<br />
chain reaction (qRT-PCR) and enzyme-linked<br />
immunosorbent assay (ELISA) showed that chemic<strong>al</strong>ly<br />
synthesized cGAMP induced IFN-b RNA<br />
and protein in L929 cells after introduction into<br />
the cells (Fig. 3A). Titration experiments showed<br />
that cGAMP induced IFN-b RNA robustly even<br />
at concentrations as low as 10 nM (Fig. 3B). Indeed,<br />
ELISA indicated that cGAMP was much<br />
more potent than c-di-GMP in inducing IFN-b<br />
(Fig. 3C). cGAMP was <strong>al</strong>so more potent than<br />
c-di-GMP and c-di-AMP in activating IRF3 (fig.<br />
S4A). To d<strong>et</strong>ermine wh<strong>et</strong>her L929 extracts contained<br />
enzymes that could synthesize other types<br />
of dinucleotides or oligonucleotides capable of<br />
activating IRF3, we tested <strong>al</strong>l four ribonucleotides<br />
in various combinations (fig. S4B). ATP<br />
and GTP were both necessary and sufficient to<br />
support the synthesis of an activator of IRF3,<br />
further supporting the idea that L929 cells contain<br />
an enzyme that synthesizes cGAMP from ATP<br />
and GTP.<br />
To d<strong>et</strong>ermine wh<strong>et</strong>her DNA virus infection<br />
leads to the production of cGAMP in cells, we<br />
A B<br />
C<br />
10 2<br />
10<br />
IFNβ RNA (fold) 103<br />
0<br />
RNAi: GFP STING<br />
Time(hr): 0 2 4 0 2 4<br />
ISD<br />
cGAMP<br />
c-di-GMP<br />
poly(I:C)<br />
IB: STING<br />
IB: Tubulin<br />
293T<br />
293T-STING<br />
DNA cGAMP DNA cGAMP DNA cGAMP<br />
HEK293T HEK293T-STING L929<br />
IB: IRF3<br />
RNAi: GFP STING<br />
IB: STING<br />
IB: Tubulin<br />
Lane: 1 2<br />
(IRF3)2<br />
IRF3<br />
(IRF3)2<br />
IRF3<br />
(IRF3)2<br />
IRF3<br />
(IRF3)2<br />
IRF3<br />
D<br />
Cold<br />
Comp<strong>et</strong>itor<br />
STING:<br />
STING/<br />
[ 32 P]-cGAMP<br />
Endogenous<br />
IRF3<br />
infected L929 cells with HSV-1 lacking ICP34.5,<br />
a vir<strong>al</strong> protein known to antagonize interferon<br />
production in the infected cells (11). Like DNA<br />
transfection, HSV-1DICP34.5 infection led to<br />
IRF3 activation in L929 cells (Fig. 3D, upper).<br />
Cell extracts from the DNA-transfected or virusinfected<br />
cells contained a heat-resistant factor that<br />
could activate IRF3 in permeabilized Raw264.7<br />
cells (Fig. 3D, lower panel). As a control, we infected<br />
L929 cells with a vesicular stomatitis virus<br />
strain, VSV = DM51[fusedtogreenfluorescent<br />
protein (VSV-DM51-GFP)], an RNA virus known<br />
to trigger strong interferon production through<br />
the RIG-I pathway (12, 13). In contrast to HSV-1,<br />
VSV-infected cells did not contain the heatresistant<br />
IRF3 activator in the same in vitro assay,<br />
<strong>al</strong>though VSV infection did induce IRF3<br />
activation in L929 cells (Fig. 3D). The heatresistant<br />
factor in HSV-1–infected cells was<br />
enriched by reversed-phase high-performance<br />
liquid chromatography (HPLC) and quantified<br />
by nano-LC-MS using SRM. DNA-transfected<br />
or HSV-1–infected cells, but not mock-treated<br />
or VSV-infected cells, produced elevated levels<br />
of cGAMP (Fig. 3E). Kin<strong>et</strong>ic experiments showed<br />
that after DNA was transfected into L929 cells,<br />
cGAMP was produced before IRF3 dimerization<br />
and IFN-b induction could be d<strong>et</strong>ected (Fig.<br />
3F). To test wh<strong>et</strong>her DNAviruses could induce<br />
cGAMP production in human cells, we infected<br />
THP1 cells with HSV1 or vaccinia virus (VACV;<br />
Fig. 3G). Both viruses induced IRF3 dimerization<br />
HEK293T<br />
Mock<br />
HSV1<br />
cGAMP<br />
Activity<br />
Assay<br />
cGAMP<br />
Mock<br />
HSV1<br />
HEK293T<br />
STING<br />
cGAMP<br />
HEK293T HEK293T<br />
STING L929<br />
Mock<br />
HSV1<br />
L929<br />
Mock<br />
HSV1<br />
cGAMP<br />
lane 1 2 3 4 5 6 7 8 9<br />
Mock<br />
HSV1<br />
Mock<br />
HSV1<br />
cGAMP<br />
lane 1 2 3 4 5 6 7<br />
[ 32 P]-ATP [ 32 P]-cGAMP<br />
*<br />
Lane:<br />
cGAMP c-di-GMP c-di-AMP<br />
- + - + + + + + + + + + + + +<br />
ATP<br />
GTP<br />
REPORTS<br />
(IRF3)2<br />
IRF3<br />
(IRF3)2<br />
IRF3<br />
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15<br />
www.sciencemag.org SCIENCE VOL 339 15 FEBRUARY 2013 829<br />
Downloaded from<br />
www.sciencemag.org on February 14, 2013