et al.
et al.
et al.
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
RESEARCH ARTICLES<br />
788<br />
expression of h-cGAS RNA was very low in<br />
human embryonic kidney (HEK) 293T cells but<br />
high in the human monocytic cell line THP1<br />
(Fig. 1C). Immunoblotting further confirmed that<br />
h-cGASproteinwasexpressedinTHP1cellsbut<br />
not HEK293T cells (Fig. 1D; no mouse cGAS<br />
antibody is available y<strong>et</strong>). Thus, the expression<br />
levels of m-cGAS and h-cGAS in different cell<br />
lines correlated with the ability of these cells to<br />
produce cGAMP and induce IFN-b in response<br />
to cytosolic DNA (4, 10).<br />
Cat<strong>al</strong>ysis by cGAS triggers type I interferon<br />
production. Overexpression of m-cGAS in<br />
HEK293T, which lacks STING expression (Fig.<br />
1D), did not induce IFN-b, whereas stable expression<br />
of STING in HEK293T cells rendered<br />
these cells highly comp<strong>et</strong>ent in IFN-b induction<br />
by m-cGAS (Fig. 2A). Point mutations of<br />
the putative cat<strong>al</strong>ytic residues Gly 198 and Ser 199<br />
to <strong>al</strong>anine abolished the ability of m-cGAS to<br />
induce IFN-b. These mutations, as well as mutations<br />
of the other putative cat<strong>al</strong>ytic residues Glu 211<br />
and Asp 213 to <strong>al</strong>anine, <strong>al</strong>so abrogated the ability<br />
of m-cGAS to induce IRF3 dimerization in<br />
HEK293T-STING cells (Fig. 2B).<br />
The magnitude of IFN-b induction by c-GAS<br />
was comparable to that induced by MAVS (an<br />
adaptor protein that functions downstream of<br />
the RNA sensor RIG-I) and was higher than that<br />
induced by other putative DNA sensors, includ-<br />
80<br />
60<br />
40<br />
20<br />
IFNβ RNA (fold) 100<br />
0<br />
shGFP<br />
sh-cGAS-a<br />
sh-cGAS-b<br />
0 1 2 3 4 5 6 7 8 9<br />
Hours post DNA transfection<br />
ing DAI, IFI16, and DDX41, by sever<strong>al</strong> orders<br />
of magnitude (Fig. 2C). To d<strong>et</strong>ermine wh<strong>et</strong>her<br />
overexpression of cGAS and other putative DNA<br />
sensors led to the production of cGAMP in cells,<br />
we incubated supernatants from heat-treated cell<br />
extracts with PFO-permeabilized Raw264.7<br />
cells, followed by measurement of IRF3 dimerization<br />
(Fig. 2D, bottom). Among <strong>al</strong>l the proteins<br />
expressed in HEK293T-STING cells, only<br />
cGAS was capable of producing the cGAMP activity<br />
in the cells.<br />
To test wh<strong>et</strong>her cGAS could synthesize cGAMP<br />
in vitro, we purified wild-type and mutant FlagcGAS<br />
proteins from transfected HEK293T cells.<br />
Wild-type m-cGAS and h-cGAS, but not the<br />
cat<strong>al</strong>ytic<strong>al</strong>ly inactive mutants of cGAS, were able<br />
to produce the cGAMP activity, which stimulated<br />
IRF3 dimerization in permeabilized Raw264.7<br />
cells (fig. S3A). We found that the in vitro activities<br />
of both m-cGAS and h-cGAS were dependent<br />
on the presence of HT-DNA (Fig. 2E). To<br />
test wh<strong>et</strong>her DNA enhances IFN-b induction by<br />
cGAS in cells, we transfected different amounts<br />
of cGAS expression plasmid, with or without<br />
HT-DNA, into HEK293T-STING cells (fig. S3B).<br />
HT-DNA markedly enhanced IFN-b induction<br />
by low (10 and 50 ng) but not high (200 ng)<br />
doses of cGAS plasmid. It is possible that the<br />
transfected cGAS plasmid DNA activated the<br />
cGAS protein in the cells, resulting in IFN-b in-<br />
A B C<br />
D<br />
E<br />
shGFP sh-cGAS<br />
HSV1: 0 2 4 6 9 0 2 4 6 9 hours<br />
IB: IRF3<br />
IB: IRF3<br />
shGFP sh-cGAS<br />
SeV: 0 4 8 0 4 8 hours<br />
(IRF3)2<br />
(IRF3)2<br />
IFNβ RNA (fold)<br />
F G<br />
shGFP sh-cGAS<br />
endogenous<br />
IRF3<br />
cGAMP<br />
activity<br />
assay<br />
50<br />
40<br />
30<br />
20<br />
15<br />
10<br />
5<br />
Mock<br />
DNA<br />
shGFP<br />
sh-cGAS<br />
shSTING<br />
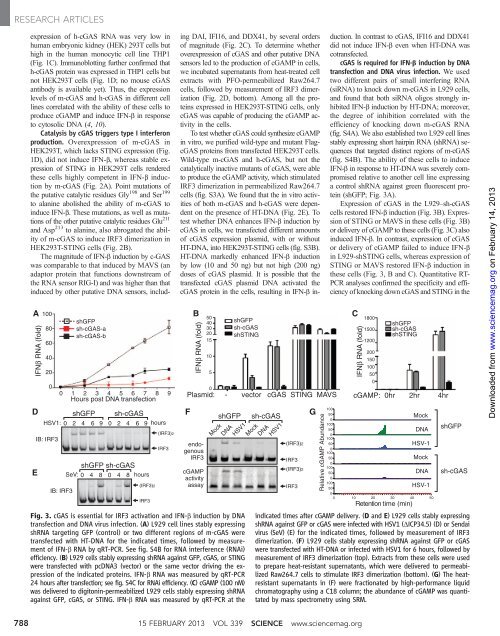
Fig. 3. cGAS is essenti<strong>al</strong> for IRF3 activation and IFN-b induction by DNA<br />
transfection and DNA virus infection. (A) L929 cell lines stably expressing<br />
shRNA targ<strong>et</strong>ing GFP (control) or two different regions of m-cGAS were<br />
transfected with HT-DNA for the indicated times, followed by measurement<br />
of IFN-b RNA by qRT-PCR. See fig. S4B for RNA interference (RNAi)<br />
efficiency. (B) L929 cells stably expressing shRNA against GFP, cGAS, or STING<br />
were transfected with pcDNA3 (vector) or the same vector driving the expression<br />
of the indicated proteins. IFN-b RNA was measured by qRT-PCR<br />
24 hours after transfection; see fig. S4C for RNAi efficiency. (C)cGAMP(100nM)<br />
was delivered to digitonin-permeabilized L929 cells stably expressing shRNA<br />
against GFP, cGAS, or STING. IFN-b RNA was measured by qRT-PCR at the<br />
IRF3<br />
IRF3<br />
0<br />
Plasmid: - vector cGAS STING MAVS<br />
HSV1<br />
Mock<br />
DNA<br />
HSV1<br />
(IRF3)2<br />
IRF3<br />
(IRF3)2<br />
IRF3<br />
Relative cGAMP Abundance<br />
duction. In contrast to cGAS, IFI16 and DDX41<br />
did not induce IFN-b even when HT-DNA was<br />
cotransfected.<br />
cGAS is required for IFN-b induction by DNA<br />
transfection and DNA virus infection. We used<br />
two different pairs of sm<strong>al</strong>l interfering RNA<br />
(siRNA) to knock down m-cGAS in L929 cells,<br />
and found that both siRNA oligos strongly inhibited<br />
IFN-b induction by HT-DNA; moreover,<br />
the degree of inhibition correlated with the<br />
efficiency of knocking down m-cGAS RNA<br />
(fig. S4A). We <strong>al</strong>so established two L929 cell lines<br />
stably expressing short hairpin RNA (shRNA) sequences<br />
that targ<strong>et</strong>ed distinct regions of m-cGAS<br />
(fig. S4B). The ability of these cells to induce<br />
IFN-b in response to HT-DNA was severely compromised<br />
relative to another cell line expressing<br />
a control shRNA against green fluorescent protein<br />
(shGFP; Fig. 3A).<br />
Expression of cGAS in the L929–sh-cGAS<br />
cells restored IFN-b induction (Fig. 3B). Expression<br />
of STING or MAVS in these cells (Fig. 3B)<br />
or delivery of cGAMP to these cells (Fig. 3C) <strong>al</strong>so<br />
induced IFN-b. In contrast, expression of cGAS<br />
or delivery of cGAMP failed to induce IFN-b<br />
in L929-shSTING cells, whereas expression of<br />
STING or MAVS restored IFN-b induction in<br />
these cells (Fig. 3, B and C). Quantitative RT-<br />
PCR an<strong>al</strong>yses confirmed the specificity and efficiency<br />
of knocking down cGAS and STING in the<br />
100<br />
50<br />
0<br />
100<br />
50<br />
0<br />
100<br />
50<br />
0<br />
100<br />
50<br />
0<br />
100<br />
50<br />
0<br />
100<br />
50<br />
0<br />
15 FEBRUARY 2013 VOL 339 SCIENCE www.sciencemag.org<br />
IFNβ RNA (fold)<br />
shGFP<br />
sh-cGAS<br />
shSTING<br />
cGAMP: 0hr 2hr 4hr<br />
Mock<br />
DNA<br />
HSV-1<br />
Mock<br />
DNA<br />
HSV-1<br />
shGFP<br />
sh-cGAS<br />
0 10 20 30 40 50<br />
R<strong>et</strong>ention time (min)<br />
indicated times after cGAMP delivery. (D and E) L929 cells stably expressing<br />
shRNA against GFP or cGAS were infected with HSV1 (DICP34.5) (D) or Sendai<br />
virus (SeV) (E) for the indicated times, followed by measurement of IRF3<br />
dimerization. (F) L929 cells stably expressing shRNA against GFP or cGAS<br />
were transfected with HT-DNA or infected with HSV1 for 6 hours, followed by<br />
measurement of IRF3 dimerization (top). Extracts from these cells were used<br />
to prepare heat-resistant supernatants, which were delivered to permeabilized<br />
Raw264.7 cells to stimulate IRF3 dimerization (bottom). (G) The heatresistant<br />
supernatants in (F) were fractionated by high-performance liquid<br />
chromatography using a C18 column; the abundance of cGAMP was quantitated<br />
by mass spectrom<strong>et</strong>ry using SRM.<br />
1800<br />
1500<br />
1200<br />
200<br />
150<br />
100<br />
50<br />
0<br />
on February 14, 2013<br />
www.sciencemag.org<br />
Downloaded from